You are browsing environment: HUMAN GUT
MGYG000002033_02135
Basic Information
help
Species
Parabacteroides massiliensis
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides massiliensis
CAZyme ID
MGYG000002033_02135
CAZy Family
GH106
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
1027
116565.2
5.4079
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000002033
4100607
MAG
Spain
Europe
Gene Location
Start: 3251;
End: 6334
Strand: -
No EC number prediction in MGYG000002033_02135.
Family
Start
End
Evalue
family coverage
GH106
30
708
8.3e-77
0.7888349514563107
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam17132
Glyco_hydro_106
1.74e-14
202
641
401
839
alpha-L-rhamnosidase.
more
pfam08531
Bac_rhamnosid_N
0.002
940
975
17
63
Alpha-L-rhamnosidase N-terminal domain. This family consists of bacterial rhamnosidase A and B enzymes. This domain is probably involved in substrate recognition.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
T2KNA8
1.73e-13
30
568
30
601
Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1
more
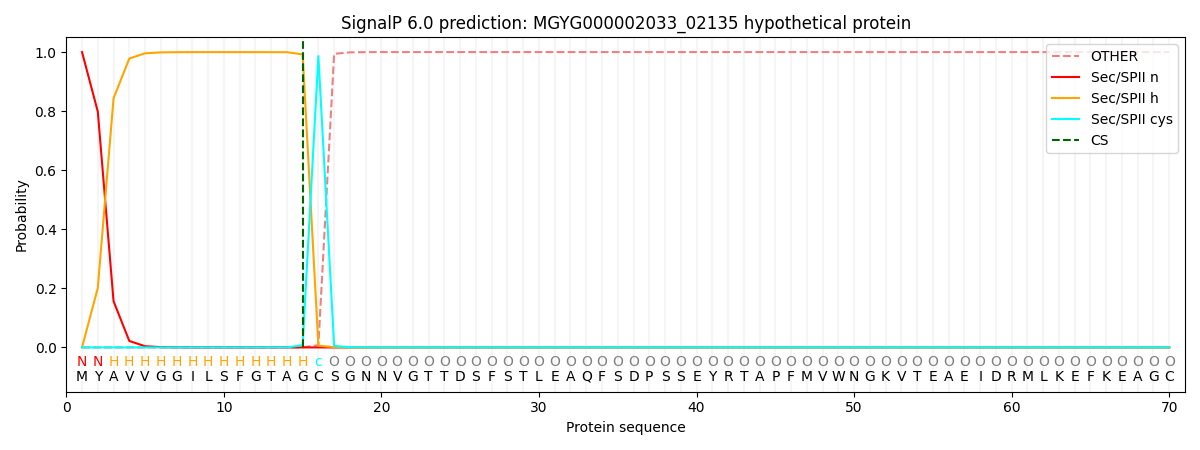
This protein is predicted as LIPO
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000002
0.000021
1.000020
0.000000
0.000000
0.000000
There is no transmembrane helices in MGYG000002033_02135.