You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002061_00598
You are here: Home > Sequence: MGYG000002061_00598
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | QAMX01 sp004555605 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; QAMX01; QAMX01; QAMX01 sp004555605 | |||||||||||
| CAZyme ID | MGYG000002061_00598 | |||||||||||
| CAZy Family | GH92 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7337; End: 9391 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH92 | 250 | 682 | 1.7e-77 | 0.9735234215885947 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07971 | Glyco_hydro_92 | 1.74e-81 | 267 | 679 | 1 | 464 | Glycosyl hydrolase family 92. Members of this family are alpha-1,2-mannosidases, enzymes which remove alpha-1,2-linked mannose residues from Man(9)(GlcNAc)(2) by hydrolysis. They are critical for the maturation of N-linked oligosaccharides and ER-associated degradation. |
| COG3537 | COG3537 | 1.23e-75 | 36 | 682 | 57 | 743 | Putative alpha-1,2-mannosidase [Carbohydrate transport and metabolism]. |
| pfam17678 | Glyco_hydro_92N | 2.81e-24 | 36 | 170 | 12 | 142 | Glycosyl hydrolase family 92 N-terminal domain. This domain is found at the N-terminus of family 92 glycosyl hydrolase proteins. |
| NF035929 | lectin_1 | 4.99e-20 | 37 | 682 | 22 | 684 | lectin. Lectins are important adhesin proteins, which bind carbohydrate structures on host cell surface. The carbohydrate specificity of diverse lectins to a large extent dictates bacteria tissue tropism by mediating specific attachment to unique host sites expressing the corresponding carbohydrate receptor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QZD53875.1 | 1.71e-131 | 2 | 682 | 7 | 725 |
| QQQ80618.1 | 1.78e-131 | 3 | 682 | 1 | 723 |
| QJW36675.1 | 4.07e-130 | 3 | 682 | 1 | 716 |
| QDV59070.1 | 6.27e-129 | 7 | 679 | 22 | 728 |
| BAK36400.1 | 1.18e-128 | 7 | 682 | 5 | 716 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6F8Z_A | 1.42e-30 | 7 | 682 | 11 | 714 | Structureof the family GH92 alpha-mannosidase BT3130 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F8Z_B Structure of the family GH92 alpha-mannosidase BT3130 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F8Z_C Structure of the family GH92 alpha-mannosidase BT3130 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F90_A Structure of the family GH92 alpha-mannosidase BT3130 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) [Bacteroides thetaiotaomicron],6F90_B Structure of the family GH92 alpha-mannosidase BT3130 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) [Bacteroides thetaiotaomicron],6F90_C Structure of the family GH92 alpha-mannosidase BT3130 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) [Bacteroides thetaiotaomicron] |
| 6DWO_A | 1.86e-19 | 344 | 682 | 312 | 697 | Crystalstructure of alpha-1-2-mannosidase from Enterococcus faecalis V583 [Enterococcus faecalis V583],6DWO_B Crystal structure of alpha-1-2-mannosidase from Enterococcus faecalis V583 [Enterococcus faecalis V583],6DWO_C Crystal structure of alpha-1-2-mannosidase from Enterococcus faecalis V583 [Enterococcus faecalis V583],6DWO_D Crystal structure of alpha-1-2-mannosidase from Enterococcus faecalis V583 [Enterococcus faecalis V583] |
| 7FE1_A | 1.86e-19 | 344 | 682 | 312 | 697 | ChainA, Alpha-1,2-mannosidase [Enterococcus faecalis ATCC 10100],7FE1_B Chain B, Alpha-1,2-mannosidase [Enterococcus faecalis ATCC 10100],7FE1_C Chain C, Alpha-1,2-mannosidase [Enterococcus faecalis ATCC 10100],7FE1_D Chain D, Alpha-1,2-mannosidase [Enterococcus faecalis ATCC 10100] |
| 7FE2_A | 4.28e-19 | 344 | 682 | 312 | 697 | ChainA, Alpha-1,2-mannosidase [Enterococcus faecalis ATCC 10100],7FE2_B Chain B, Alpha-1,2-mannosidase [Enterococcus faecalis ATCC 10100],7FE2_C Chain C, Alpha-1,2-mannosidase [Enterococcus faecalis ATCC 10100],7FE2_D Chain D, Alpha-1,2-mannosidase [Enterococcus faecalis ATCC 10100] |
| 6F91_A | 1.54e-16 | 36 | 682 | 21 | 726 | Structureof the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F91_B Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F91_C Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F91_D Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F91_E Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F91_F Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F91_G Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F91_H Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F92_A Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) [Bacteroides thetaiotaomicron],6F92_B Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) [Bacteroides thetaiotaomicron],6F92_C Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) [Bacteroides thetaiotaomicron],6F92_D Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D4ATR3 | 2.71e-19 | 36 | 681 | 44 | 743 | Uncharacterized secreted glycosidase ARB_07629 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07629 PE=1 SV=1 |
| O86365 | 6.43e-18 | 37 | 682 | 62 | 753 | Uncharacterized glycosidase Rv0584 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=Rv0584 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
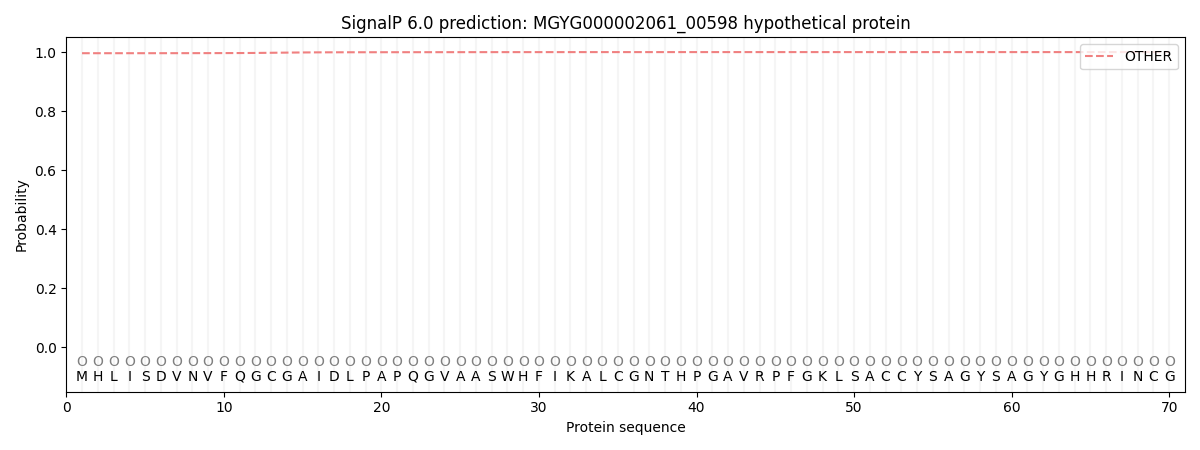
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.996621 | 0.003343 | 0.000051 | 0.000007 | 0.000003 | 0.000006 |
