You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002063_00848
You are here: Home > Sequence: MGYG000002063_00848
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; | |||||||||||
| CAZyme ID | MGYG000002063_00848 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3032; End: 6058 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 624 | 942 | 3.5e-92 | 0.9735973597359736 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 6.77e-102 | 621 | 941 | 1 | 308 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 2.04e-96 | 666 | 939 | 1 | 261 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 2.02e-71 | 617 | 939 | 31 | 335 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| PRK15319 | PRK15319 | 2.19e-04 | 310 | 473 | 665 | 844 | fibronectin-binding autotransporter adhesin ShdA. |
| COG4625 | COG4625 | 0.002 | 329 | 454 | 35 | 160 | Uncharacterized conserved protein, contains a C-terminal beta-barrel porin domain [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADX05746.1 | 2.38e-98 | 678 | 981 | 7 | 310 |
| ADX05712.1 | 4.09e-72 | 618 | 920 | 69 | 380 |
| QBI56661.1 | 1.30e-68 | 618 | 945 | 46 | 359 |
| AOS63897.1 | 1.14e-67 | 620 | 938 | 48 | 350 |
| SHI05302.1 | 2.24e-65 | 618 | 945 | 37 | 348 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1XYZ_A | 2.45e-70 | 621 | 938 | 28 | 337 | ChainA, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus],1XYZ_B Chain B, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus] |
| 6FHE_A | 1.39e-61 | 632 | 941 | 23 | 338 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 3WUF_A | 1.40e-59 | 652 | 945 | 33 | 312 | Themutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 [Streptomyces sp.],3WUG_A The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
| 3WUB_A | 1.73e-58 | 652 | 945 | 33 | 312 | Thewild type crystal structure of b-1,4-Xylanase (XynAS9) from Streptomyces sp. 9 [Streptomyces sp.],3WUE_A The wild type crystal structure of b-1,4-Xylanase (XynAS9) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
| 1GOK_A | 1.17e-57 | 650 | 947 | 33 | 303 | Thermostablexylanase I from Thermoascus aurantiacus- Crystal form II [Thermoascus aurantiacus],1GOM_A Thermostable xylanase I from Thermoascus aurantiacus- Crystal form I [Thermoascus aurantiacus],1GOO_A Thermostable xylanase I from Thermoascus aurantiacus - Cryocooled glycerol complex [Thermoascus aurantiacus],1GOQ_A Thermostable xylanase I from Thermoascus aurantiacus - Room temperature xylobiose complex [Thermoascus aurantiacus],1GOR_A THERMOSTABLE XYLANASE I FROM THERMOASCUS AURANTIACUS - XYLOBIOSE COMPLEX AT 100 K [Thermoascus aurantiacus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10478 | 5.28e-65 | 621 | 938 | 518 | 827 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
| Q0H904 | 3.81e-58 | 622 | 945 | 32 | 325 | Endo-1,4-beta-xylanase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=xlnC PE=2 SV=2 |
| B4XVN1 | 5.62e-57 | 620 | 945 | 35 | 350 | Endo-1,4-beta-xylanase A OS=Streptomyces sp. OX=1931 GN=xynAS9 PE=1 SV=1 |
| B0Y6E0 | 9.30e-57 | 622 | 945 | 32 | 316 | Probable endo-1,4-beta-xylanase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=xlnC PE=2 SV=2 |
| P23360 | 1.35e-56 | 650 | 947 | 59 | 329 | Endo-1,4-beta-xylanase OS=Thermoascus aurantiacus OX=5087 GN=XYNA PE=1 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as SP
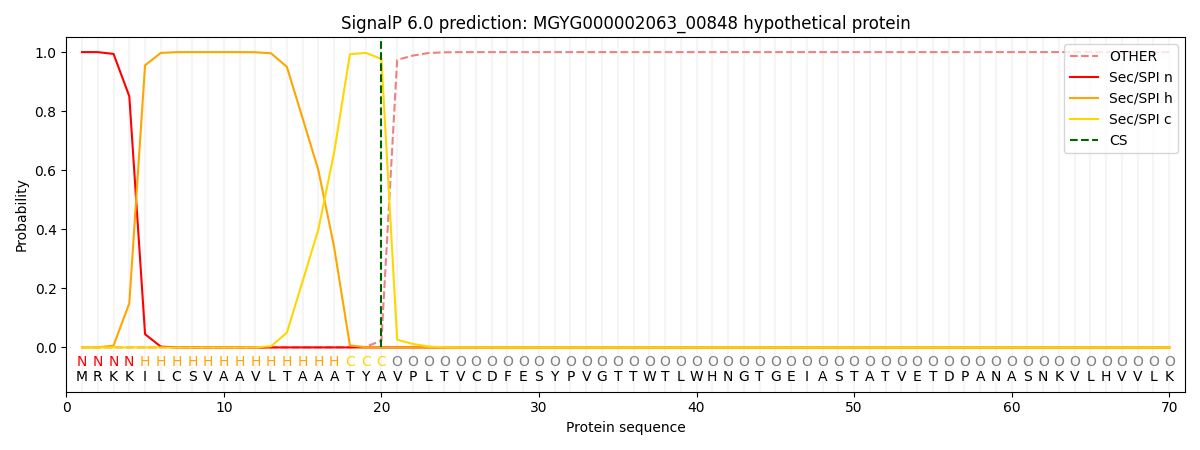
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000349 | 0.998883 | 0.000192 | 0.000207 | 0.000183 | 0.000161 |
