You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002075_01098
You are here: Home > Sequence: MGYG000002075_01098
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
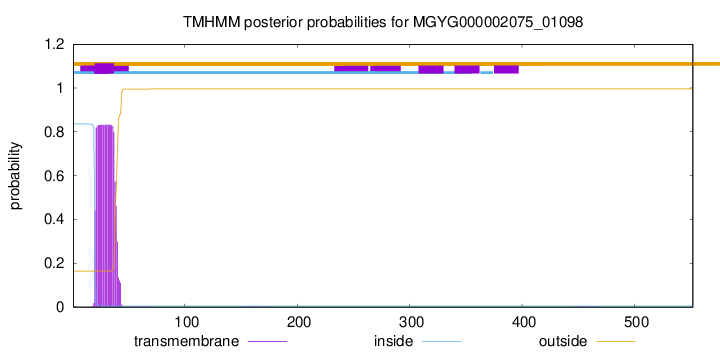
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; ; | |||||||||||
| CAZyme ID | MGYG000002075_01098 | |||||||||||
| CAZy Family | GH77 | |||||||||||
| CAZyme Description | 4-alpha-glucanotransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4245; End: 5903 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH77 | 25 | 506 | 1.7e-172 | 0.9919028340080972 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK14508 | PRK14508 | 0.0 | 16 | 510 | 2 | 497 | 4-alpha-glucanotransferase; Provisional |
| pfam02446 | Glyco_hydro_77 | 0.0 | 25 | 486 | 1 | 456 | 4-alpha-glucanotransferase. These enzymes EC:2.4.1.25 transfer a segment of a (1,4)-alpha-D-glucan to a new 4-position in an acceptor, which may be glucose or (1,4)-alpha-D-glucan. |
| PLN02635 | PLN02635 | 2.22e-169 | 16 | 525 | 25 | 538 | disproportionating enzyme |
| COG1640 | MalQ | 8.81e-168 | 7 | 510 | 2 | 515 | 4-alpha-glucanotransferase [Carbohydrate transport and metabolism]. |
| TIGR00217 | malQ | 3.40e-130 | 18 | 486 | 13 | 490 | 4-alpha-glucanotransferase. This enzyme is known as amylomaltase and disproportionating enzyme. [Energy metabolism, Biosynthesis and degradation of polysaccharides] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIX92984.1 | 1.25e-235 | 17 | 504 | 1 | 491 |
| QJU21270.1 | 4.98e-235 | 17 | 504 | 20 | 510 |
| QQR02334.1 | 1.03e-234 | 17 | 504 | 11 | 501 |
| ANU48762.2 | 1.03e-234 | 17 | 504 | 11 | 501 |
| QRP42403.1 | 2.20e-234 | 17 | 504 | 3 | 493 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1CWY_A | 2.85e-153 | 17 | 505 | 3 | 495 | CrystalStructure Of Amylomaltase From Thermus Aquaticus, A Glycosyltransferase Catalysing The Production Of Large Cyclic Glucans [Thermus aquaticus],1ESW_A X-Ray Structure Of Acarbose Bound To Amylomaltase From Thermus Aquaticus. Implications For The Synthesis Of Large Cyclic Glucans [Thermus aquaticus] |
| 1FP8_A | 1.14e-152 | 17 | 505 | 3 | 495 | StructureOf The Amylomaltase From Thermus Thermophilus Hb8 In Space Group P21212 [Thermus thermophilus],1FP9_A Structure Of Amylomaltase From Thermus Thermophilus Hb8 In Space Group C2 [Thermus thermophilus] |
| 2OWC_A | 1.39e-151 | 12 | 505 | 1 | 497 | Structureof a covalent intermediate in Thermus thermophilus amylomaltase [Thermus thermophilus],2OWW_A Covalent intermediate in amylomaltase in complex with the acceptor analog 4-deoxyglucose [Thermus thermophilus],2OWX_A THERMUS THERMOPHILUS AMYLOMALTASE AT pH 5.6 [Thermus thermophilus] |
| 5JIW_A | 2.60e-151 | 17 | 505 | 3 | 495 | Crystalstructure of Thermus aquaticus amylomaltase (GH77) in complex with a 34-meric cycloamylose [Thermus aquaticus] |
| 2X1I_A | 1.08e-147 | 20 | 505 | 6 | 495 | glycosidehydrolase family 77 4-alpha-glucanotransferase from thermus brockianus [Thermus brockianus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0A3Q1 | 1.90e-165 | 18 | 504 | 3 | 490 | 4-alpha-glucanotransferase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=malQ PE=3 SV=1 |
| P0A3Q0 | 1.90e-165 | 18 | 504 | 3 | 490 | 4-alpha-glucanotransferase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=malQ PE=3 SV=1 |
| Q59266 | 1.18e-164 | 27 | 511 | 3 | 487 | 4-alpha-glucanotransferase OS=Clostridium butyricum OX=1492 GN=malQ PE=1 SV=1 |
| O87172 | 1.56e-152 | 17 | 505 | 3 | 495 | 4-alpha-glucanotransferase OS=Thermus thermophilus OX=274 GN=malQ PE=1 SV=1 |
| P72785 | 1.05e-140 | 18 | 505 | 4 | 495 | 4-alpha-glucanotransferase OS=Synechocystis sp. (strain PCC 6803 / Kazusa) OX=1111708 GN=malQ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
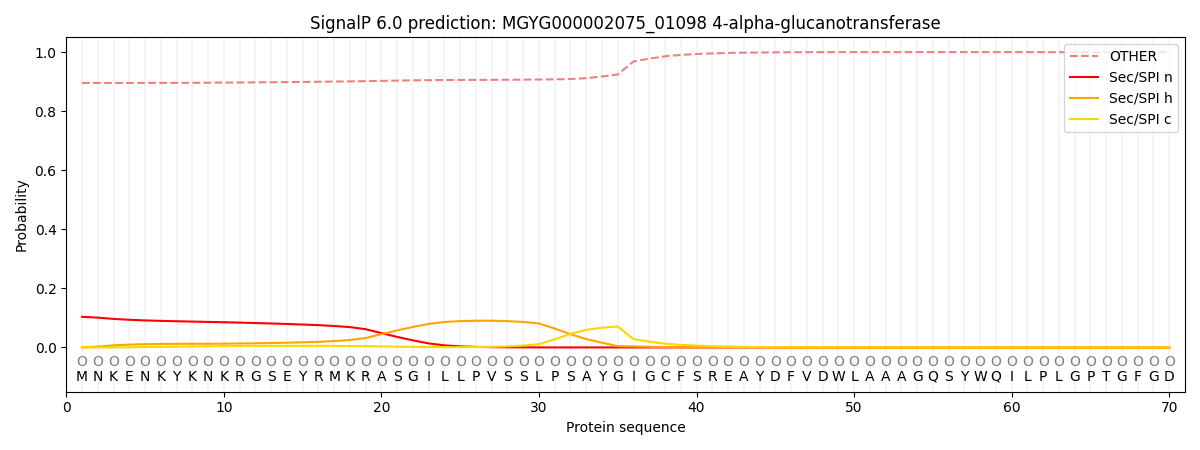
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.900321 | 0.098213 | 0.001031 | 0.000167 | 0.000106 | 0.000180 |