You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002092_00256
You are here: Home > Sequence: MGYG000002092_00256
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; | |||||||||||
| CAZyme ID | MGYG000002092_00256 | |||||||||||
| CAZy Family | GH105 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6896; End: 8884 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03629 | SASA | 1.18e-07 | 426 | 545 | 102 | 224 | Carbohydrate esterase, sialic acid-specific acetylesterase. The catalytic triad of this esterase enzyme comprises residues Ser127, His403 and Asp391 in UniProtKB:P70665. |
| pfam02837 | Glyco_hydro_2_N | 3.70e-05 | 277 | 368 | 38 | 134 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| pfam03629 | SASA | 5.15e-04 | 180 | 216 | 65 | 102 | Carbohydrate esterase, sialic acid-specific acetylesterase. The catalytic triad of this esterase enzyme comprises residues Ser127, His403 and Asp391 in UniProtKB:P70665. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCO21319.1 | 1.06e-131 | 4 | 662 | 3 | 643 |
| CCO21108.1 | 1.06e-131 | 4 | 662 | 3 | 643 |
| CCO21289.1 | 1.06e-131 | 4 | 662 | 3 | 643 |
| CCO21210.1 | 1.06e-131 | 4 | 662 | 3 | 643 |
| CCO21153.1 | 1.06e-131 | 4 | 662 | 3 | 643 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7KMM_A | 9.21e-133 | 28 | 655 | 28 | 636 | ChainA, Sialic acid-specific 9-O-acetylesterase [Xanthomonas citri pv. citri str. 306],7KMM_B Chain B, Sialic acid-specific 9-O-acetylesterase [Xanthomonas citri pv. citri str. 306] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P82450 | 2.05e-32 | 29 | 650 | 31 | 533 | Sialate O-acetylesterase OS=Rattus norvegicus OX=10116 GN=Siae PE=1 SV=2 |
| P70665 | 7.60e-30 | 29 | 650 | 31 | 532 | Sialate O-acetylesterase OS=Mus musculus OX=10090 GN=Siae PE=1 SV=3 |
| Q9HAT2 | 3.96e-28 | 30 | 650 | 33 | 506 | Sialate O-acetylesterase OS=Homo sapiens OX=9606 GN=SIAE PE=1 SV=1 |
| Q5RFU0 | 7.40e-27 | 30 | 650 | 33 | 506 | Sialate O-acetylesterase OS=Pongo abelii OX=9601 GN=SIAE PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
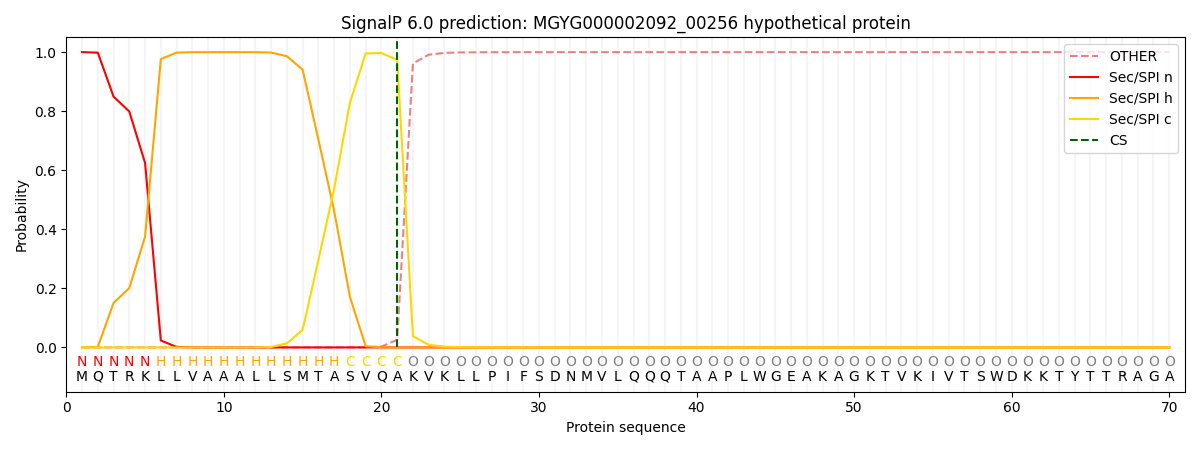
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000348 | 0.998814 | 0.000283 | 0.000194 | 0.000172 | 0.000147 |
