You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002108_00976
You are here: Home > Sequence: MGYG000002108_00976
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900290275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900290275 | |||||||||||
| CAZyme ID | MGYG000002108_00976 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | Extracellular exo-alpha-L-arabinofuranosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12685; End: 15123 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH51 | 161 | 812 | 1.8e-109 | 0.8793650793650793 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3534 | AbfA | 1.16e-45 | 203 | 812 | 33 | 499 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
| smart00813 | Alpha-L-AF_C | 3.21e-21 | 449 | 552 | 1 | 104 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| pfam06964 | Alpha-L-AF_C | 7.11e-21 | 449 | 552 | 1 | 107 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNT66893.1 | 0.0 | 2 | 812 | 3 | 813 |
| AGB28822.1 | 0.0 | 8 | 812 | 8 | 805 |
| BCS85815.1 | 0.0 | 15 | 812 | 3 | 795 |
| ADE81175.1 | 0.0 | 3 | 812 | 19 | 827 |
| AGB28798.1 | 2.68e-242 | 1 | 811 | 2 | 810 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ZPS_AAA | 8.14e-91 | 30 | 533 | 8 | 524 | ChainAAA, MgGH51 [Meripilus giganteus],6ZPV_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPW_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPX_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPY_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPZ_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ0_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ1_AAA Chain AAA, MgGH51 [Meripilus giganteus] |
| 3S2C_A | 6.57e-26 | 219 | 536 | 48 | 365 | Structureof the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_B Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_C Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_D Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_E Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_F Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_G Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_H Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_I Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_J Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_K Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_L Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1] |
| 4ATW_A | 8.62e-26 | 219 | 536 | 48 | 365 | Thecrystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_B The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_C The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_D The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_E The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_F The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8] |
| 3UG3_A | 1.06e-25 | 219 | 536 | 68 | 385 | Crystalstructure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG4_A Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG5_A Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima] |
| 1PZ3_A | 5.11e-17 | 218 | 366 | 50 | 179 | Crystalstructure of a family 51 (GH51) alpha-L-arabinofuranosidase from Geobacillus stearothermophilus T6 [Geobacillus stearothermophilus],1PZ3_B Crystal structure of a family 51 (GH51) alpha-L-arabinofuranosidase from Geobacillus stearothermophilus T6 [Geobacillus stearothermophilus],6SXU_AAA Chain AAA, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Geobacillus stearothermophilus],6SXU_BBB Chain BBB, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Geobacillus stearothermophilus],6SXV_A GH51 a-l-arabinofuranosidase soaked with aziridine inhibitor [Geobacillus stearothermophilus],6SXV_B GH51 a-l-arabinofuranosidase soaked with aziridine inhibitor [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P82593 | 2.64e-145 | 24 | 812 | 37 | 823 | Extracellular exo-alpha-L-arabinofuranosidase OS=Streptomyces chartreusis OX=1969 PE=1 SV=1 |
| Q9SG80 | 1.79e-110 | 22 | 539 | 41 | 558 | Alpha-L-arabinofuranosidase 1 OS=Arabidopsis thaliana OX=3702 GN=ASD1 PE=1 SV=1 |
| Q8VZR2 | 9.82e-101 | 19 | 533 | 37 | 547 | Alpha-L-arabinofuranosidase 2 OS=Arabidopsis thaliana OX=3702 GN=ASD2 PE=2 SV=1 |
| Q0CTV2 | 4.57e-90 | 22 | 541 | 28 | 540 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=abfA PE=3 SV=1 |
| U6A629 | 2.66e-88 | 8 | 527 | 7 | 524 | Alpha-L-arabinofuranosidase A OS=Penicillium canescens OX=5083 GN=abfA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
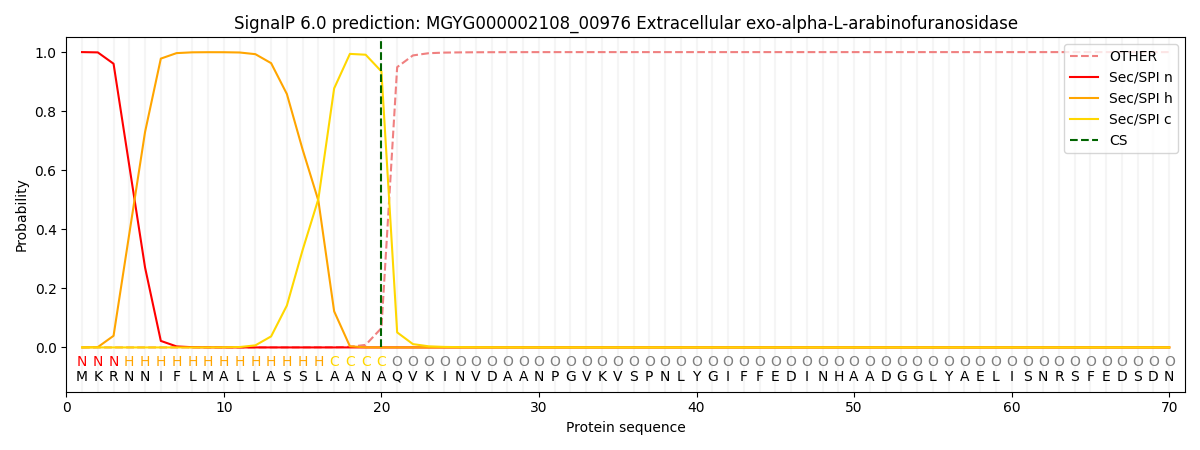
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000427 | 0.998775 | 0.000219 | 0.000213 | 0.000180 | 0.000170 |
