You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002132_00030
You are here: Home > Sequence: MGYG000002132_00030
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
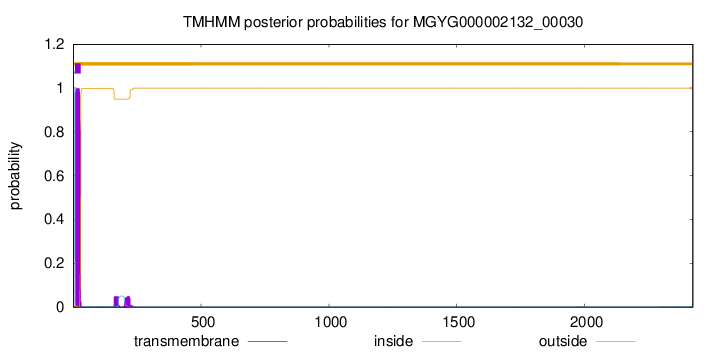
TMHMM annotations
Basic Information help
| Species | Monoglobus sp900542675 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales; Monoglobaceae; Monoglobus; Monoglobus sp900542675 | |||||||||||
| CAZyme ID | MGYG000002132_00030 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 48459; End: 55733 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02773 | PLN02773 | 3.95e-22 | 719 | 1016 | 34 | 263 | pectinesterase |
| PLN02682 | PLN02682 | 1.57e-19 | 632 | 1057 | 15 | 366 | pectinesterase family protein |
| PLN02432 | PLN02432 | 5.84e-17 | 679 | 1016 | 10 | 253 | putative pectinesterase |
| pfam01095 | Pectinesterase | 1.31e-16 | 696 | 1017 | 11 | 260 | Pectinesterase. |
| PLN02170 | PLN02170 | 9.18e-15 | 662 | 1059 | 205 | 518 | probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUO19783.1 | 0.0 | 1 | 2419 | 1 | 2730 |
| AUO19003.1 | 3.96e-127 | 1 | 397 | 1 | 398 |
| AUO19782.1 | 1.78e-124 | 1 | 385 | 1 | 384 |
| QUT74167.1 | 4.32e-77 | 2072 | 2419 | 46 | 396 |
| QCD41145.1 | 1.45e-74 | 2066 | 2419 | 33 | 392 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5C1C_A | 5.47e-12 | 862 | 1021 | 114 | 264 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
| 5C1E_A | 5.47e-12 | 862 | 1021 | 114 | 264 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
| 2NSP_A | 2.04e-11 | 695 | 1026 | 16 | 292 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 1GQ8_A | 9.37e-11 | 866 | 1057 | 114 | 307 | Pectinmethylesterase from Carrot [Daucus carota] |
| 4PMH_A | 4.26e-10 | 866 | 1013 | 178 | 338 | Thestructure of rice weevil pectin methyl esterase [Sitophilus oryzae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9LVQ0 | 1.68e-15 | 792 | 1016 | 88 | 263 | Pectinesterase 31 OS=Arabidopsis thaliana OX=3702 GN=PME31 PE=1 SV=1 |
| P38536 | 3.49e-13 | 30 | 202 | 1681 | 1856 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38535 | 6.61e-13 | 30 | 202 | 907 | 1082 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| Q7Y201 | 5.14e-12 | 866 | 1053 | 407 | 598 | Probable pectinesterase/pectinesterase inhibitor 13 OS=Arabidopsis thaliana OX=3702 GN=PME13 PE=2 SV=2 |
| O22149 | 5.32e-12 | 866 | 1062 | 308 | 505 | Probable pectinesterase/pectinesterase inhibitor 17 OS=Arabidopsis thaliana OX=3702 GN=PME17 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
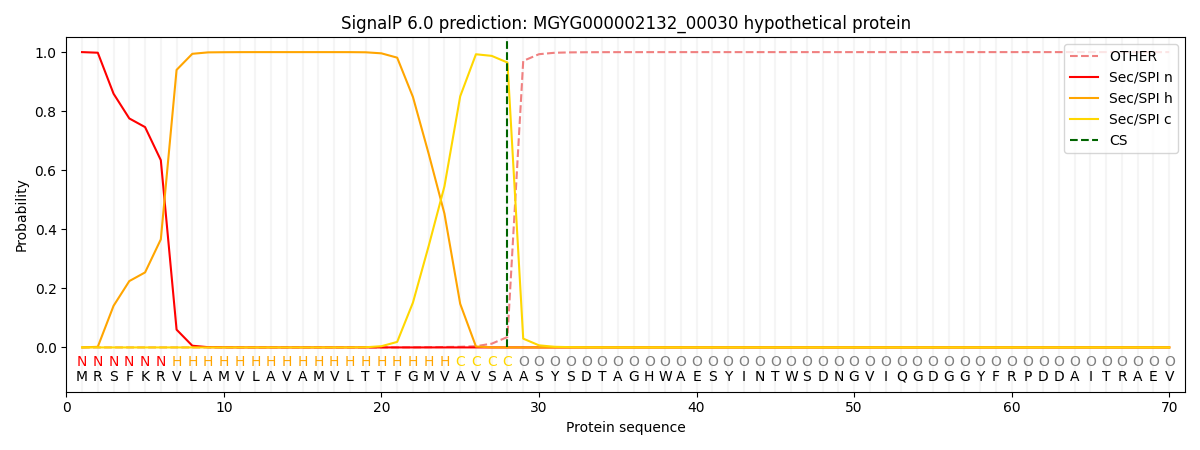
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000253 | 0.999057 | 0.000175 | 0.000176 | 0.000158 | 0.000138 |