You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002132_00133
You are here: Home > Sequence: MGYG000002132_00133
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
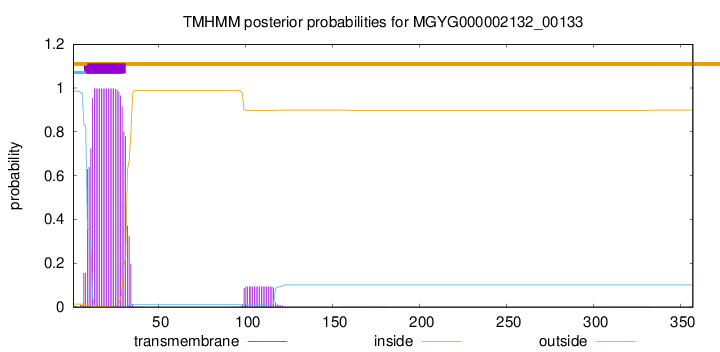
TMHMM annotations
Basic Information help
| Species | Monoglobus sp900542675 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales; Monoglobaceae; Monoglobus; Monoglobus sp900542675 | |||||||||||
| CAZyme ID | MGYG000002132_00133 | |||||||||||
| CAZy Family | SLH | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 52197; End: 53270 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| SLH | 33 | 73 | 9.5e-18 | 0.9761904761904762 |
| SLH | 150 | 191 | 2.8e-17 | 0.9523809523809523 |
| SLH | 93 | 132 | 1.6e-16 | 0.9285714285714286 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033190 | inl_like_NEAT_1 | 6.22e-17 | 38 | 196 | 588 | 748 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam09479 | Flg_new | 1.09e-16 | 252 | 319 | 1 | 65 | Listeria-Bacteroides repeat domain (List_Bact_rpt). This model describes a conserved core region of about 43 residues, which occurs in at least two families of tandem repeats. These include 78-residue repeats which occur from 2 to 15 times in some proteins of Bacteroides forsythus ATCC 43037, and 70-residue repeats found in families of internalins of Listeria species. Single copies are found in proteins of Fibrobacter succinogenes, Geobacter sulfurreducens, and a few other bacteria. |
| pfam00395 | SLH | 3.76e-11 | 93 | 132 | 3 | 42 | S-layer homology domain. |
| pfam00395 | SLH | 3.95e-11 | 150 | 191 | 1 | 41 | S-layer homology domain. |
| pfam00395 | SLH | 2.10e-10 | 33 | 73 | 1 | 42 | S-layer homology domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAU26536.1 | 5.90e-46 | 35 | 198 | 34 | 197 |
| AUO19782.1 | 8.04e-43 | 3 | 197 | 2 | 198 |
| QGQ95647.1 | 1.81e-41 | 27 | 198 | 27 | 201 |
| BAM67143.1 | 7.74e-41 | 3 | 204 | 10 | 208 |
| AUO19783.1 | 2.95e-40 | 3 | 318 | 2 | 378 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6BT4_A | 8.79e-11 | 39 | 201 | 33 | 197 | Crystalstructure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit [Bacillus anthracis] |
| 3PYW_A | 8.93e-11 | 39 | 201 | 12 | 176 | Thestructure of the SLH domain from B. anthracis surface array protein at 1.8A [Bacillus anthracis] |
| 4AQ1_A | 4.11e-06 | 42 | 196 | 12 | 164 | Structureof the SbsB S-layer protein of Geobacillus stearothermophilus PV72p2 in complex with nanobody KB6 [Geobacillus stearothermophilus] |
| 4AQ1_C | 4.11e-06 | 42 | 196 | 12 | 164 | Structureof the SbsB S-layer protein of Geobacillus stearothermophilus PV72p2 in complex with nanobody KB6 [Geobacillus stearothermophilus] |
| 2Y5P_A | 5.58e-06 | 252 | 318 | 5 | 68 | B-repeatof Listeria monocytogenes InlB (internalin B) [Listeria monocytogenes EGD],2Y5P_B B-repeat of Listeria monocytogenes InlB (internalin B) [Listeria monocytogenes EGD],2Y5P_C B-repeat of Listeria monocytogenes InlB (internalin B) [Listeria monocytogenes EGD],2Y5P_D B-repeat of Listeria monocytogenes InlB (internalin B) [Listeria monocytogenes EGD] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38536 | 8.00e-20 | 33 | 196 | 1682 | 1851 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38537 | 1.31e-19 | 33 | 199 | 36 | 205 | Surface-layer 125 kDa protein OS=Lysinibacillus sphaericus OX=1421 PE=3 SV=1 |
| P38535 | 7.51e-19 | 33 | 196 | 908 | 1077 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| P19424 | 2.32e-18 | 2 | 196 | 3 | 210 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| C6CRV0 | 2.06e-17 | 40 | 196 | 1291 | 1455 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
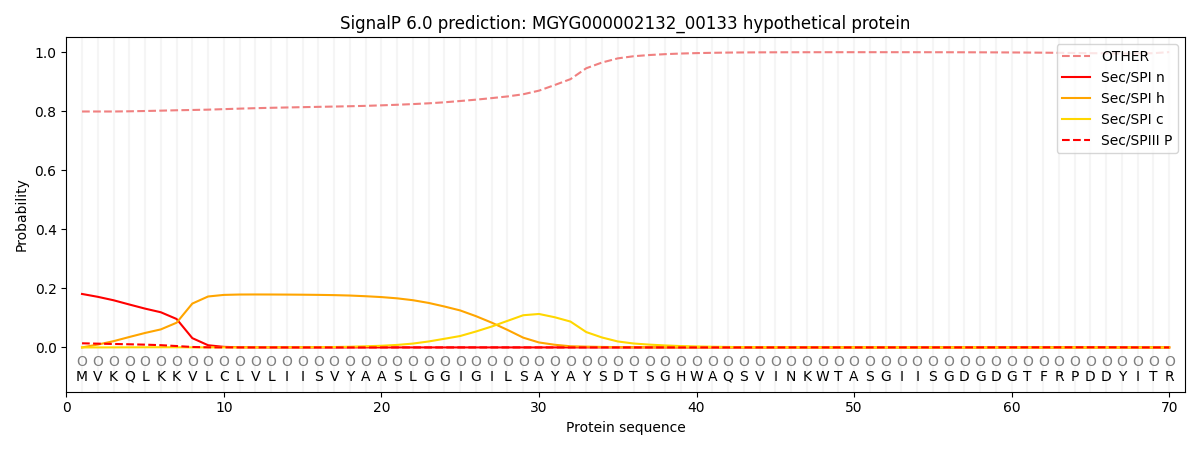
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.808260 | 0.167686 | 0.007291 | 0.000885 | 0.000524 | 0.015364 |