You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002134_02307
You are here: Home > Sequence: MGYG000002134_02307
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Mailhella sp900541395 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Desulfobacterota; Desulfovibrionia; Desulfovibrionales; Desulfovibrionaceae; Mailhella; Mailhella sp900541395 | |||||||||||
| CAZyme ID | MGYG000002134_02307 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1207; End: 2319 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 104 | 321 | 2.1e-45 | 0.9444444444444444 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 2.82e-60 | 38 | 367 | 7 | 313 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PRK05337 | PRK05337 | 6.27e-59 | 41 | 326 | 3 | 282 | beta-hexosaminidase; Provisional |
| pfam00933 | Glyco_hydro_3 | 1.88e-52 | 39 | 365 | 7 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK15098 | PRK15098 | 9.86e-09 | 154 | 351 | 147 | 338 | beta-glucosidase BglX. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMD90486.1 | 3.89e-108 | 29 | 365 | 38 | 383 |
| AAS96712.1 | 5.97e-108 | 29 | 369 | 51 | 394 |
| ABM28024.1 | 5.97e-108 | 29 | 369 | 51 | 394 |
| ADP87229.1 | 5.97e-108 | 29 | 369 | 51 | 394 |
| EGJ48598.1 | 8.19e-103 | 6 | 369 | 10 | 375 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6K5J_A | 1.46e-49 | 39 | 367 | 18 | 338 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
| 4GVF_A | 2.14e-37 | 42 | 326 | 4 | 281 | Crystalstructure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) bound to GlcNAc [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4GVF_B Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) bound to GlcNAc [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4GVG_A Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4GVG_B Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4GVH_A Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) covalently bound to 5-fluoro-GlcNAc. [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4GVH_B Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) covalently bound to 5-fluoro-GlcNAc. [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4HZM_A Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) bound to N-[(3S,4R,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]butanamide [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4HZM_B Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) bound to N-[(3S,4R,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]butanamide [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 5G1M_A | 4.43e-37 | 42 | 344 | 25 | 311 | Crystalstructure of NagZ from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G1M_B Crystal structure of NagZ from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G2M_A Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine [Pseudomonas aeruginosa PAO1],5G2M_B Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine [Pseudomonas aeruginosa PAO1],5G3R_A Crystal Structure Of Nagz From Pseudomonas Aeruginosa In Complex With N-acetylglucosamine And L-ala-1,6-anhydromurnac [Pseudomonas aeruginosa PAO1],5G3R_B Crystal Structure Of Nagz From Pseudomonas Aeruginosa In Complex With N-acetylglucosamine And L-ala-1,6-anhydromurnac [Pseudomonas aeruginosa PAO1],5G5K_A Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1],5G5K_B Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1] |
| 4GVI_A | 1.12e-36 | 42 | 326 | 4 | 281 | Crystalstructure of mutant (D248N) Salmonella typhimurium family 3 glycoside hydrolase (NagZ) in complex with GlcNAc-1,6-anhMurNAc [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4GVI_B Crystal structure of mutant (D248N) Salmonella typhimurium family 3 glycoside hydrolase (NagZ) in complex with GlcNAc-1,6-anhMurNAc [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 3TEV_A | 1.63e-36 | 32 | 365 | 13 | 327 | Thecrystal structure of glycosyl hydrolase from Deinococcus radiodurans R1 [Deinococcus radiodurans R1],3TEV_B The crystal structure of glycosyl hydrolase from Deinococcus radiodurans R1 [Deinococcus radiodurans R1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q87BR5 | 1.71e-44 | 41 | 355 | 1 | 314 | Beta-hexosaminidase OS=Xylella fastidiosa (strain Temecula1 / ATCC 700964) OX=183190 GN=nagZ PE=3 SV=1 |
| B2I6G9 | 1.71e-44 | 41 | 355 | 1 | 314 | Beta-hexosaminidase OS=Xylella fastidiosa (strain M23) OX=405441 GN=nagZ PE=3 SV=1 |
| Q0A911 | 4.79e-43 | 42 | 326 | 6 | 285 | Beta-hexosaminidase OS=Alkalilimnicola ehrlichii (strain ATCC BAA-1101 / DSM 17681 / MLHE-1) OX=187272 GN=nagZ PE=3 SV=1 |
| B2FPW9 | 9.70e-43 | 41 | 355 | 1 | 314 | Beta-hexosaminidase OS=Stenotrophomonas maltophilia (strain K279a) OX=522373 GN=nagZ PE=3 SV=1 |
| Q9PAZ0 | 9.70e-43 | 41 | 355 | 1 | 314 | Beta-hexosaminidase OS=Xylella fastidiosa (strain 9a5c) OX=160492 GN=nagZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
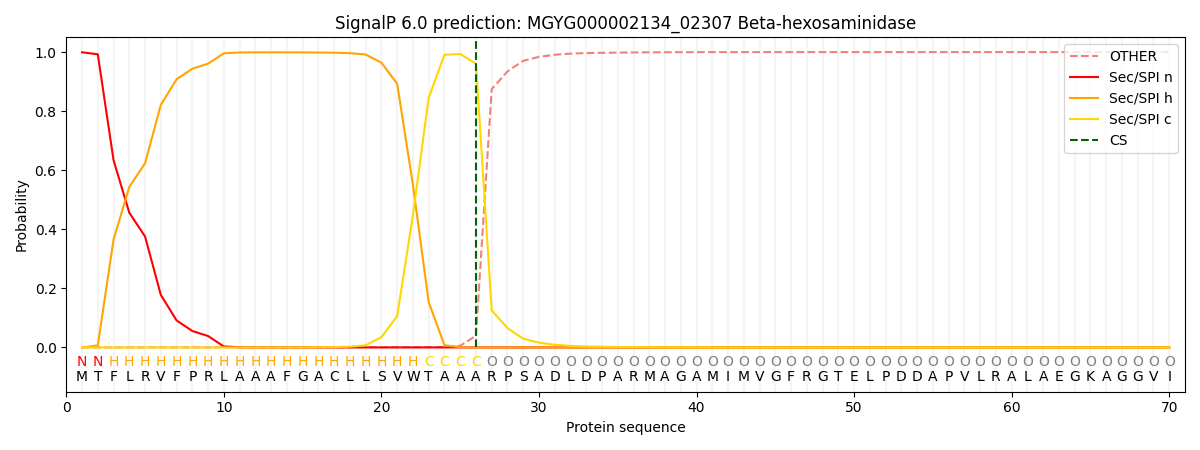
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000983 | 0.997576 | 0.000837 | 0.000220 | 0.000180 | 0.000178 |
