You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002148_01637
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Eisenbergiella;
|
| CAZyme ID |
MGYG000002148_01637
|
| CAZy Family |
GT83 |
| CAZyme Description |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 498 |
|
58010.24 |
7.4038 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000002148 |
4597814 |
MAG |
United States |
North America |
|
| Gene Location |
Start: 40081;
End: 41577
Strand: -
|
No EC number prediction in MGYG000002148_01637.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
19 |
383 |
2.2e-60 |
0.6481481481481481 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1807
|
ArnT |
7.21e-27 |
16 |
354 |
8 |
335 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231
|
PMT_2 |
6.36e-18 |
75 |
231 |
2 |
160 |
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| PRK13279
|
arnT |
1.44e-05 |
16 |
332 |
7 |
310 |
lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| COG5305
|
COG5305 |
0.006 |
75 |
351 |
97 |
390 |
Uncharacterized membrane protein [Function unknown]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
O67270
|
1.32e-08 |
43 |
385 |
29 |
351 |
Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1 |
This protein is predicted as OTHER
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.994195
|
0.002330
|
0.000291
|
0.000011
|
0.000008
|
0.003197
|
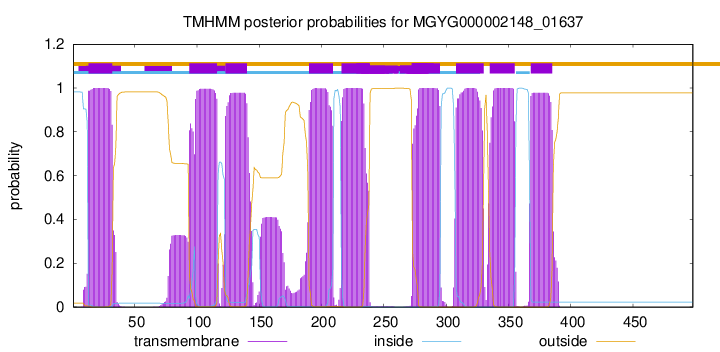
| start |
end |
| 13 |
32 |
| 94 |
116 |
| 123 |
140 |
| 190 |
209 |
| 216 |
238 |
| 273 |
295 |
| 308 |
330 |
| 335 |
355 |
| 368 |
385 |


