You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002197_00199
You are here: Home > Sequence: MGYG000002197_00199
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
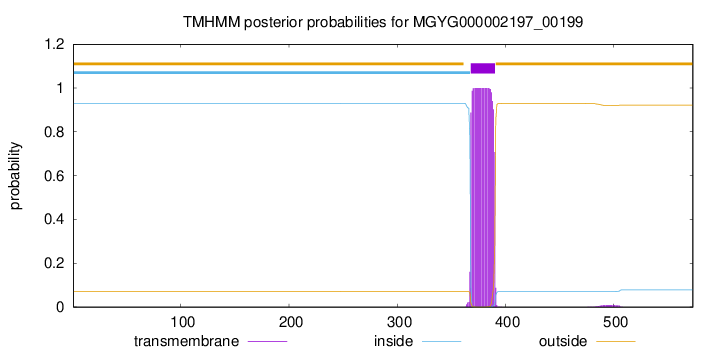
TMHMM annotations
Basic Information help
| Species | Parvimonas sp001553085 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Tissierellales; Peptoniphilaceae; Parvimonas; Parvimonas sp001553085 | |||||||||||
| CAZyme ID | MGYG000002197_00199 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | Dephospho-CoA kinase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 29049; End: 30770 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 412 | 537 | 7.4e-26 | 0.7925925925925926 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd16896 | LT_Slt70-like | 4.32e-64 | 395 | 541 | 1 | 146 | uncharacterized lytic transglycosylase subfamily with similarity to Slt70. Uncharacterized lytic transglycosylase (LT) with a conserved sequence pattern suggesting similarity to the Slt70, a 70kda soluble lytic transglycosylase which also has an N-terminal U-shaped U-domain and a linker L-domain. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| cd13401 | Slt70-like | 4.10e-44 | 394 | 532 | 2 | 142 | 70kDa soluble lytic transglycosylase (Slt70) and similar proteins. Catalytic domain of the 70kda soluble lytic transglycosylase (LT)-like proteins, which also have an N-terminal U-shaped U-domain and a linker L-domain. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria and the LTs in bacteriophage lambda. |
| PRK00081 | coaE | 1.19e-43 | 5 | 199 | 1 | 192 | dephospho-CoA kinase; Reviewed |
| COG0237 | CoaE | 3.92e-41 | 4 | 195 | 1 | 187 | Dephospho-CoA kinase [Coenzyme transport and metabolism]. |
| cd02022 | DPCK | 8.86e-41 | 8 | 189 | 2 | 179 | Dephospho-coenzyme A kinase (DPCK, EC 2.7.1.24) catalyzes the phosphorylation of dephosphocoenzyme A (dCoA) to yield CoA, which is the final step in CoA biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AXU10287.1 | 9.77e-246 | 1 | 573 | 1 | 569 |
| VEH94495.1 | 5.95e-230 | 1 | 573 | 1 | 569 |
| AIZ37242.1 | 7.06e-108 | 390 | 573 | 29 | 212 |
| QQK07141.1 | 8.67e-43 | 390 | 544 | 29 | 183 |
| AVQ45663.1 | 4.54e-39 | 394 | 543 | 28 | 179 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1SLY_A | 7.03e-19 | 394 | 543 | 449 | 598 | ComplexOf The 70-Kda Soluble Lytic Transglycosylase With Bulgecin A [Escherichia coli] |
| 1QSA_A | 2.88e-18 | 394 | 543 | 449 | 598 | CrystalStructure Of The 70 Kda Soluble Lytic Transglycosylase Slt70 From Escherichia Coli At 1.65 Angstroms Resolution [Escherichia coli],1QTE_A Crystal Structure Of The 70 Kda Soluble Lytic Transglycosylase Slt70 From Escherichia Coli At 1.90 A Resolution In Complex With A 1,6- Anhydromurotripeptide [Escherichia coli] |
| 6FBT_A | 3.38e-16 | 394 | 546 | 445 | 595 | ChainA, Lytic murein transglycosylase [Pseudomonas aeruginosa] |
| 5OHU_A | 3.57e-16 | 394 | 546 | 474 | 624 | TheX-ray Structure of Lytic Transglycosylase Slt from Pseudomonas aeruginosa [Pseudomonas aeruginosa] |
| 2IF2_A | 3.79e-16 | 6 | 199 | 2 | 190 | CrystalStructure of the Putative Dephospho-CoA Kinase from Aquifex aeolicus, Northeast Structural Genomics Target QR72. [Aquifex aeolicus VF5],2IF2_B Crystal Structure of the Putative Dephospho-CoA Kinase from Aquifex aeolicus, Northeast Structural Genomics Target QR72. [Aquifex aeolicus VF5],2IF2_C Crystal Structure of the Putative Dephospho-CoA Kinase from Aquifex aeolicus, Northeast Structural Genomics Target QR72. [Aquifex aeolicus VF5] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8RHR7 | 5.92e-24 | 8 | 199 | 6 | 191 | Dephospho-CoA kinase OS=Fusobacterium nucleatum subsp. nucleatum (strain ATCC 25586 / DSM 15643 / BCRC 10681 / CIP 101130 / JCM 8532 / KCTC 2640 / LMG 13131 / VPI 4355) OX=190304 GN=coaE PE=3 SV=1 |
| O34932 | 1.46e-22 | 8 | 195 | 5 | 190 | Dephospho-CoA kinase OS=Bacillus subtilis (strain 168) OX=224308 GN=coaE PE=3 SV=1 |
| Q8DSZ0 | 1.14e-20 | 6 | 195 | 3 | 189 | Dephospho-CoA kinase OS=Streptococcus mutans serotype c (strain ATCC 700610 / UA159) OX=210007 GN=coaE PE=3 SV=1 |
| Q3ABL6 | 1.38e-20 | 8 | 194 | 7 | 191 | Dephospho-CoA kinase OS=Carboxydothermus hydrogenoformans (strain ATCC BAA-161 / DSM 6008 / Z-2901) OX=246194 GN=coaE PE=3 SV=1 |
| Q5M575 | 2.81e-20 | 8 | 194 | 3 | 186 | Dephospho-CoA kinase OS=Streptococcus thermophilus (strain ATCC BAA-250 / LMG 18311) OX=264199 GN=coaE PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
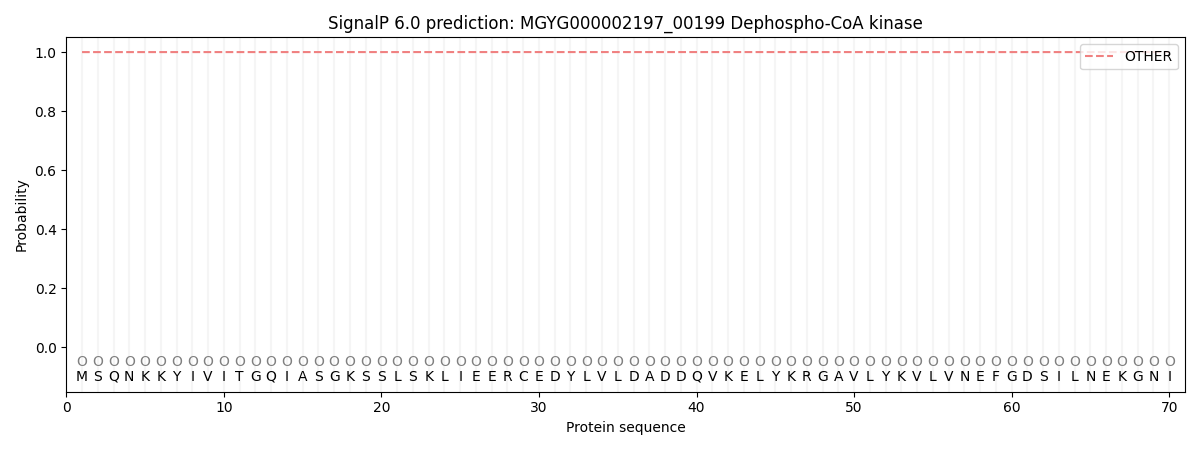
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000075 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |