You are browsing environment: HUMAN GUT
MGYG000002209_01535
Basic Information
help
Species
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; CAG-462;
CAZyme ID
MGYG000002209_01535
CAZy Family
GH139
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000002209
3647221
MAG
Spain
Europe
Gene Location
Start: 12818;
End: 15169
Strand: -
Family
Start
End
Evalue
family coverage
GH139
20
775
0
0.9794079794079794
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam18961
DUF5703_N
1.05e-157
42
322
2
287
Domain of unknown function (DUF5703). This is an N-terminal domain of unknown function mostly found in bacteria. It is possible that this domain might be a putative glycoside hydrolase. This family belongs to the Galactose Mutarotase-like superfamily.
more
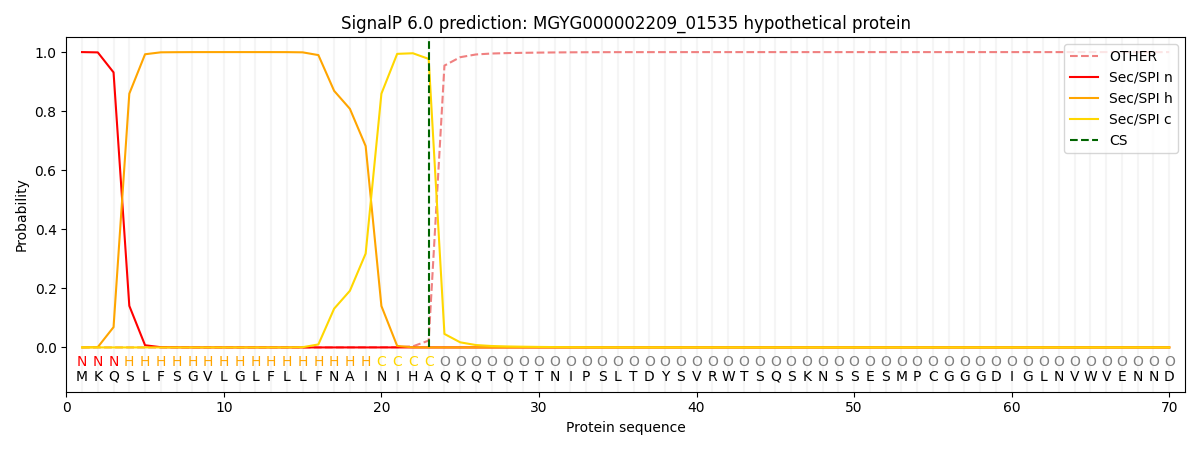
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000282
0.999052
0.000172
0.000176
0.000155
0.000150
There is no transmembrane helices in MGYG000002209_01535.