You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002216_00629
You are here: Home > Sequence: MGYG000002216_00629
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; ; | |||||||||||
| CAZyme ID | MGYG000002216_00629 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12248; End: 15628 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 517 | 710 | 1.7e-35 | 0.9351851851851852 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 5.58e-26 | 47 | 380 | 387 | 760 | beta-glucosidase BglX. |
| pfam14310 | Fn3-like | 6.59e-19 | 304 | 375 | 1 | 71 | Fibronectin type III-like domain. This domain has a fibronectin type III-like structure. It is often found in association with pfam00933 and pfam01915. Its function is unknown. |
| pfam01915 | Glyco_hydro_3_C | 4.58e-14 | 55 | 270 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| pfam00933 | Glyco_hydro_3 | 5.35e-13 | 534 | 750 | 87 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| cd14256 | Dockerin_I | 1.06e-11 | 1062 | 1118 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCZ48953.1 | 1.13e-187 | 32 | 710 | 34 | 806 |
| AQR97225.1 | 3.35e-180 | 1 | 733 | 1 | 830 |
| AGF58529.1 | 7.67e-178 | 1 | 733 | 1 | 830 |
| QUN34102.1 | 5.60e-159 | 29 | 733 | 37 | 927 |
| ABR35983.1 | 2.69e-158 | 29 | 733 | 37 | 927 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5WUG_A | 3.22e-86 | 44 | 753 | 37 | 821 | Expression,characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii],5WVP_A Expression, characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii] |
| 2X42_A | 2.13e-47 | 44 | 386 | 325 | 713 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with alpha-D-glucose [Thermotoga neapolitana DSM 4359] |
| 2X40_A | 2.13e-47 | 44 | 386 | 325 | 713 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with glycerol [Thermotoga neapolitana DSM 4359],2X41_A Structure of beta-glucosidase 3B from Thermotoga neapolitana in complex with glucose [Thermotoga neapolitana DSM 4359] |
| 5WAB_A | 8.64e-25 | 45 | 387 | 306 | 664 | CrystalStructure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_B Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_C Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_D Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703] |
| 3ABZ_A | 4.40e-22 | 208 | 388 | 645 | 838 | Crystalstructure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_B Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_C Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_D Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16084 | 2.50e-72 | 22 | 732 | 10 | 809 | Beta-glucosidase A OS=Butyrivibrio fibrisolvens OX=831 GN=bglA PE=3 SV=1 |
| P15885 | 4.05e-69 | 47 | 754 | 13 | 765 | Beta-glucosidase OS=Ruminococcus albus OX=1264 PE=3 SV=1 |
| F6C6C1 | 3.01e-25 | 513 | 754 | 49 | 288 | Exo-alpha-(1->6)-L-arabinopyranosidase OS=Bifidobacterium breve (strain ACS-071-V-Sch8b) OX=866777 GN=HMPREF9228_1477 PE=1 SV=1 |
| E7CY69 | 1.59e-24 | 513 | 754 | 49 | 288 | Exo-alpha-(1->6)-L-arabinopyranosidase OS=Bifidobacterium longum OX=216816 GN=apy PE=1 SV=1 |
| Q5BFG8 | 5.15e-23 | 535 | 709 | 69 | 247 | Beta-glucosidase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
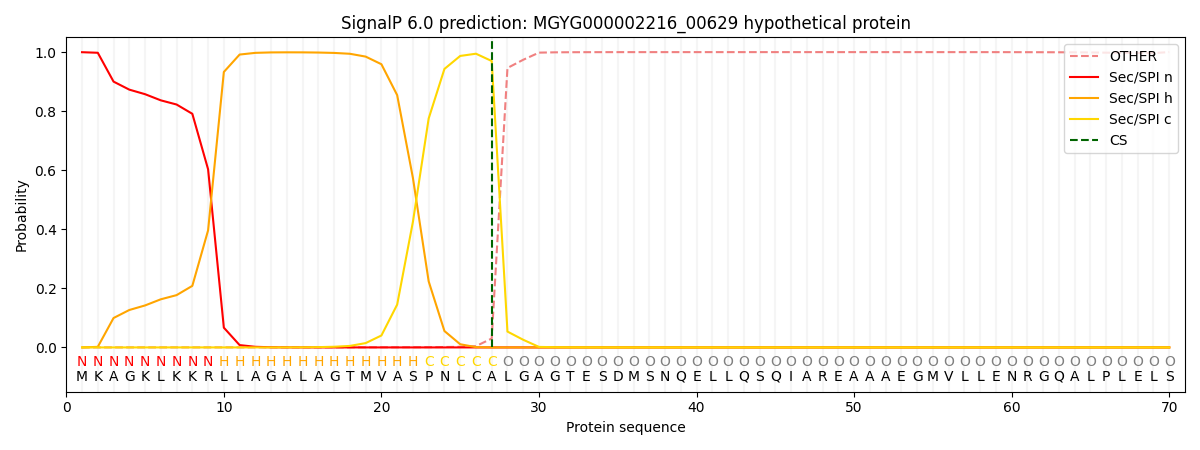
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002883 | 0.994334 | 0.000921 | 0.000954 | 0.000517 | 0.000331 |
