You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002218_00580
You are here: Home > Sequence: MGYG000002218_00580
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp000436495 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp000436495 | |||||||||||
| CAZyme ID | MGYG000002218_00580 | |||||||||||
| CAZy Family | GH35 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1941; End: 4280 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 39 | 350 | 1.1e-116 | 0.990228013029316 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01301 | Glyco_hydro_35 | 5.09e-163 | 38 | 350 | 1 | 315 | Glycosyl hydrolases family 35. |
| PLN03059 | PLN03059 | 2.29e-53 | 33 | 609 | 31 | 726 | beta-galactosidase; Provisional |
| COG1874 | GanA | 1.84e-41 | 37 | 606 | 6 | 600 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| pfam00754 | F5_F8_type_C | 3.47e-07 | 702 | 772 | 2 | 70 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam13364 | BetaGal_dom4_5 | 0.003 | 504 | 598 | 19 | 109 | Beta-galactosidase jelly roll domain. This domain is found in beta galactosidase enzymes. It has a jelly roll fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT50204.1 | 0.0 | 1 | 778 | 1 | 779 |
| QRO23434.1 | 0.0 | 17 | 777 | 19 | 780 |
| QUU04522.1 | 0.0 | 30 | 779 | 19 | 769 |
| QKH86407.1 | 0.0 | 30 | 779 | 19 | 769 |
| AUI47952.1 | 0.0 | 30 | 779 | 19 | 769 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6EON_A | 0.0 | 12 | 777 | 11 | 777 | GalactanaseBT0290 [Bacteroides thetaiotaomicron VPI-5482] |
| 3D3A_A | 2.15e-302 | 28 | 628 | 4 | 605 | Crystalstructure of a beta-galactosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 4MAD_A | 7.89e-141 | 38 | 596 | 23 | 578 | ChainA, Beta-galactosidase [Niallia circulans],4MAD_B Chain B, Beta-galactosidase [Niallia circulans] |
| 4E8C_A | 7.15e-122 | 39 | 618 | 10 | 593 | Crystalstructure of streptococcal beta-galactosidase in complex with galactose [Streptococcus pneumoniae TIGR4],4E8C_B Crystal structure of streptococcal beta-galactosidase in complex with galactose [Streptococcus pneumoniae TIGR4],4E8D_A Crystal structure of streptococcal beta-galactosidase [Streptococcus pneumoniae TIGR4],4E8D_B Crystal structure of streptococcal beta-galactosidase [Streptococcus pneumoniae TIGR4] |
| 3WF3_A | 1.87e-117 | 25 | 596 | 26 | 622 | Crystalstructure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_B Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_C Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_D Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF4_A Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_B Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_C Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_D Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P48982 | 8.85e-132 | 39 | 579 | 37 | 573 | Beta-galactosidase OS=Xanthomonas manihotis OX=43353 GN=bga PE=1 SV=1 |
| Q60HF6 | 1.95e-118 | 12 | 612 | 11 | 644 | Beta-galactosidase OS=Macaca fascicularis OX=9541 GN=GLB1 PE=2 SV=1 |
| Q9TRY9 | 1.43e-117 | 11 | 612 | 11 | 645 | Beta-galactosidase OS=Canis lupus familiaris OX=9615 GN=GLB1 PE=1 SV=3 |
| P16278 | 2.57e-117 | 12 | 596 | 12 | 621 | Beta-galactosidase OS=Homo sapiens OX=9606 GN=GLB1 PE=1 SV=2 |
| Q58D55 | 2.16e-115 | 12 | 611 | 11 | 642 | Beta-galactosidase OS=Bos taurus OX=9913 GN=GLB1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
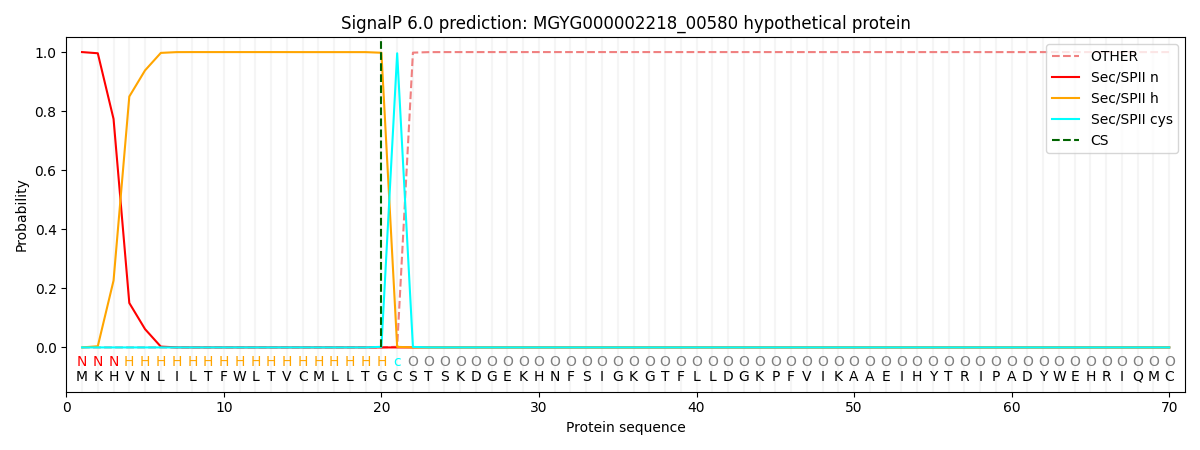
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000052 | 0.000000 | 0.000000 | 0.000000 |
