You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002218_02163
You are here: Home > Sequence: MGYG000002218_02163
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
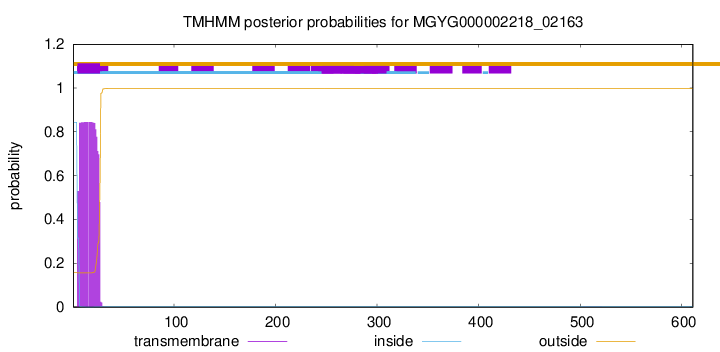
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp000436495 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp000436495 | |||||||||||
| CAZyme ID | MGYG000002218_02163 | |||||||||||
| CAZy Family | GH110 | |||||||||||
| CAZyme Description | Alpha-1,3-galactosidase B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 24379; End: 26214 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH110 | 30 | 596 | 2e-221 | 0.9981751824817519 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT50242.1 | 9.77e-288 | 17 | 608 | 14 | 598 |
| EDN88509.1 | 7.54e-285 | 17 | 608 | 14 | 598 |
| QUT76583.1 | 3.91e-271 | 30 | 594 | 27 | 595 |
| QUT21221.1 | 1.37e-268 | 7 | 611 | 4 | 606 |
| QIX64824.1 | 2.76e-268 | 7 | 611 | 4 | 606 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JW4_A | 2.32e-77 | 24 | 602 | 21 | 601 | Crystalstructure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta],7JW4_B Crystal structure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta] |
| 7JWF_A | 1.23e-76 | 24 | 602 | 21 | 601 | Crystalstructure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_B Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_C Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_D Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A6LFT2 | 7.37e-267 | 7 | 611 | 4 | 606 | Alpha-1,3-galactosidase B OS=Parabacteroides distasonis (strain ATCC 8503 / DSM 20701 / CIP 104284 / JCM 5825 / NCTC 11152) OX=435591 GN=glaB PE=3 SV=1 |
| Q64XV2 | 9.30e-266 | 4 | 595 | 2 | 595 | Alpha-1,3-galactosidase B OS=Bacteroides fragilis (strain YCH46) OX=295405 GN=glaB PE=3 SV=1 |
| Q5LGZ8 | 1.32e-265 | 4 | 595 | 2 | 595 | Alpha-1,3-galactosidase B OS=Bacteroides fragilis (strain ATCC 25285 / DSM 2151 / CCUG 4856 / JCM 11019 / NCTC 9343 / Onslow) OX=272559 GN=glaB PE=1 SV=1 |
| A6KWM0 | 1.94e-264 | 15 | 595 | 9 | 592 | Alpha-1,3-galactosidase B OS=Phocaeicola vulgatus (strain ATCC 8482 / DSM 1447 / JCM 5826 / CCUG 4940 / NBRC 14291 / NCTC 11154) OX=435590 GN=glaB1 PE=3 SV=1 |
| A6L2M8 | 3.37e-261 | 27 | 601 | 20 | 598 | Alpha-1,3-galactosidase B OS=Phocaeicola vulgatus (strain ATCC 8482 / DSM 1447 / JCM 5826 / CCUG 4940 / NBRC 14291 / NCTC 11154) OX=435590 GN=glaB2 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
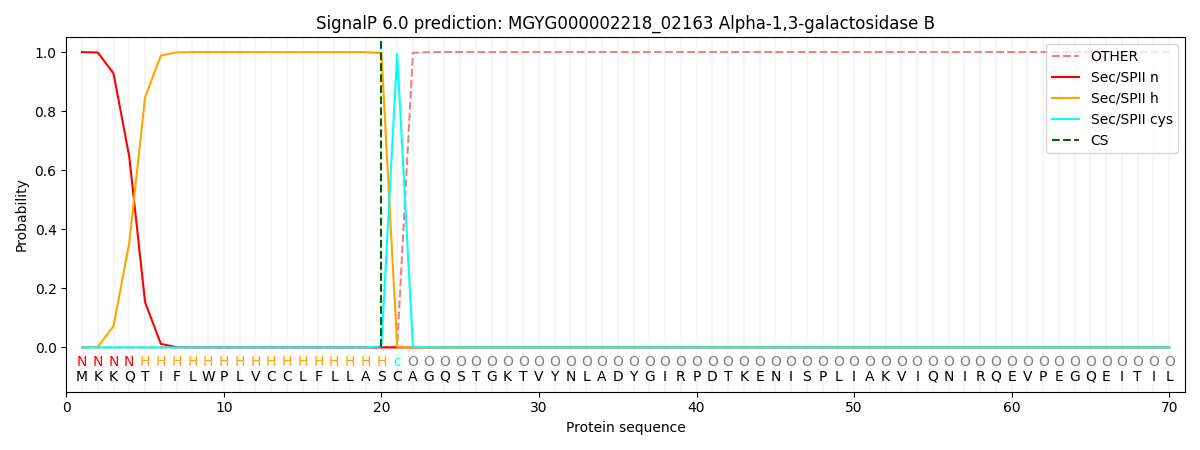
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000049 | 0.000000 | 0.000000 | 0.000000 |