You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002219_00225
You are here: Home > Sequence: MGYG000002219_00225
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Kiritimatiellae; RFP12; UBA1067; UBA1731; | |||||||||||
| CAZyme ID | MGYG000002219_00225 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Glycosyl hydrolase family 109 protein 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2095; End: 3537 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 49 | 450 | 8.7e-153 | 0.9874686716791979 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 1.42e-14 | 63 | 373 | 19 | 300 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 9.56e-09 | 63 | 174 | 15 | 117 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| PRK11579 | PRK11579 | 0.006 | 114 | 254 | 57 | 187 | putative oxidoreductase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AIM37953.1 | 5.39e-135 | 45 | 449 | 52 | 447 |
| QQD11965.1 | 1.40e-132 | 45 | 456 | 52 | 454 |
| QLE88973.1 | 5.93e-132 | 4 | 450 | 5 | 448 |
| QLE81030.1 | 5.93e-132 | 4 | 450 | 5 | 448 |
| AVR47122.1 | 6.39e-132 | 3 | 472 | 5 | 465 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6T2B_A | 1.83e-100 | 39 | 450 | 32 | 440 | Glycosidehydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_B Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_C Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_D Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila] |
| 2IXA_A | 7.79e-65 | 51 | 454 | 21 | 437 | A-zyme,N-acetylgalactosaminidase [Elizabethkingia meningoseptica],2IXB_A Crystal structure of N-ACETYLGALACTOSAMINIDASE in complex with GalNAC [Elizabethkingia meningoseptica] |
| 3KUX_A | 5.75e-06 | 56 | 253 | 8 | 189 | Structureof the YPO2259 putative oxidoreductase from Yersinia pestis [Yersinia pestis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0HTG8 | 2.32e-124 | 4 | 450 | 5 | 448 | Glycosyl hydrolase family 109 protein 2 OS=Shewanella sp. (strain MR-7) OX=60481 GN=Shewmr7_2602 PE=3 SV=1 |
| A0KYQ9 | 6.58e-124 | 4 | 450 | 5 | 448 | Glycosyl hydrolase family 109 protein 2 OS=Shewanella sp. (strain ANA-3) OX=94122 GN=Shewana3_2701 PE=3 SV=1 |
| Q0HH61 | 9.30e-124 | 4 | 450 | 5 | 448 | Glycosyl hydrolase family 109 protein 2 OS=Shewanella sp. (strain MR-4) OX=60480 GN=Shewmr4_2535 PE=3 SV=1 |
| A8H2K3 | 1.86e-123 | 1 | 450 | 1 | 447 | Glycosyl hydrolase family 109 protein OS=Shewanella pealeana (strain ATCC 700345 / ANG-SQ1) OX=398579 GN=Spea_1465 PE=3 SV=1 |
| B1KNF7 | 2.71e-120 | 46 | 450 | 52 | 458 | Glycosyl hydrolase family 109 protein OS=Shewanella woodyi (strain ATCC 51908 / MS32) OX=392500 GN=Swoo_1749 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TATLIPO
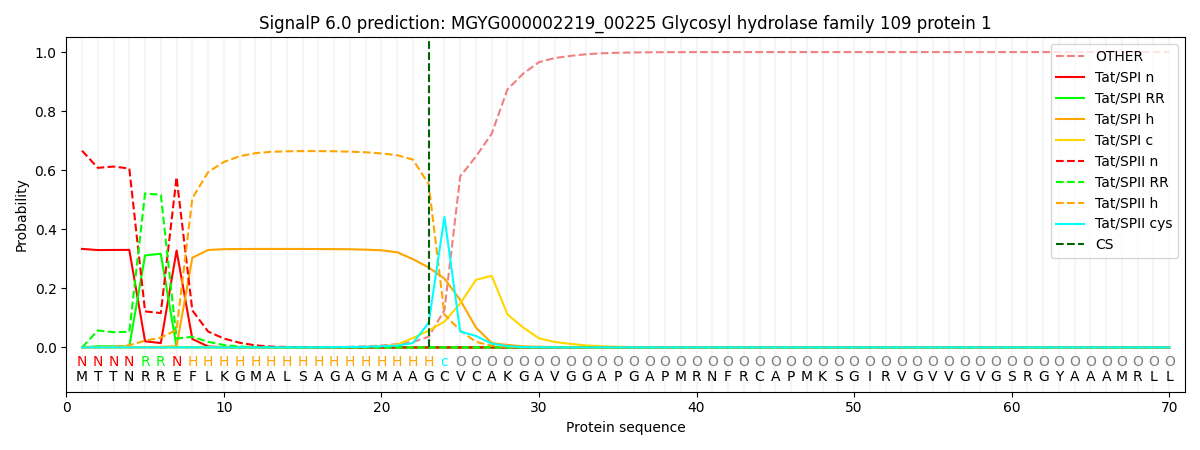
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000013 | 0.000005 | 0.000242 | 0.333589 | 0.666152 | 0.000000 |
