You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002230_00325
You are here: Home > Sequence: MGYG000002230_00325
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ruminococcus sp900545125 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus; Ruminococcus sp900545125 | |||||||||||
| CAZyme ID | MGYG000002230_00325 | |||||||||||
| CAZy Family | GH11 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16535; End: 20437 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 454 | 781 | 2.3e-90 | 0.9933993399339934 |
| GH11 | 53 | 247 | 1e-56 | 0.9887005649717514 |
| CBM22 | 280 | 407 | 3.2e-35 | 0.9694656488549618 |
| CBM22 | 922 | 1046 | 8.2e-30 | 0.9541984732824428 |
| CE4 | 1092 | 1207 | 1.5e-22 | 0.8461538461538461 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 3.08e-96 | 455 | 781 | 1 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 1.42e-85 | 496 | 779 | 1 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 4.15e-70 | 472 | 784 | 43 | 342 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| cd10954 | CE4_CtAXE_like | 1.09e-65 | 1095 | 1282 | 1 | 180 | Catalytic NodB homology domain of Clostridium thermocellum acetylxylan esterase and its bacterial homologs. This family is represented by Clostridium thermocellum acetylxylan esterase (CtAXE, EC 3.1.1.72), a member of the carbohydrate esterase 4 (CE4) superfamily. CtAXE deacetylates O-acetylated xylan, a key component of plant cell walls. It shows no detectable activity on generic esterase substrates including para-nitrophenyl acetate. It is specific for sugar-based substrates and will precipitate acetylxylan, as a consequence of deacetylation. CtAXE is a monomeric protein containing a catalytic NodB homology domain with the same overall topology and a deformed (beta/alpha)8 barrel fold as other CE4 esterases. However, due to differences in the topography of the substrate-binding groove, the chemistry of the active center, and metal ion coordination, CtAXE has different metal ion preference and lacks activity on N-acetyl substrates. It is significantly activated by Co2+. Moreover, CtAXE displays distinctly different ligand coordination to the metal ion, utilizing an aspartate, a histidine, and four water molecules, as opposed to the conserved His-His-Asp zinc-binding triad of other CE4 esterases. |
| pfam00457 | Glyco_hydro_11 | 3.06e-48 | 53 | 246 | 1 | 175 | Glycosyl hydrolases family 11. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADU24192.1 | 1.36e-225 | 29 | 782 | 33 | 785 |
| CBL18305.1 | 1.28e-183 | 17 | 1300 | 16 | 883 |
| CAL91982.1 | 1.37e-154 | 453 | 783 | 9 | 335 |
| CDG46726.1 | 6.86e-147 | 452 | 782 | 25 | 351 |
| CAA77476.1 | 2.06e-146 | 452 | 783 | 627 | 953 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2C1F_A | 9.17e-131 | 38 | 259 | 2 | 223 | Thestructure of the family 11 xylanase from Neocallimastix patriciarum [Neocallimastix patriciarum] |
| 2VG9_A | 6.00e-125 | 38 | 254 | 2 | 216 | Crystalstructure of Loop Swap mutant of Necallimastix patriciarum Xyn11A [Neocallimastix patriciarum] |
| 2W5F_A | 9.85e-97 | 275 | 782 | 21 | 527 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2W5F_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 2WYS_A | 1.57e-93 | 337 | 782 | 83 | 527 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WYS_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_A Chain A, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 6FHE_A | 9.76e-56 | 455 | 780 | 13 | 339 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P29126 | 4.12e-147 | 452 | 783 | 627 | 953 | Bifunctional endo-1,4-beta-xylanase XylA OS=Ruminococcus flavefaciens OX=1265 GN=xynA PE=3 SV=1 |
| P29127 | 1.17e-127 | 14 | 270 | 6 | 263 | Bifunctional endo-1,4-beta-xylanase A OS=Neocallimastix patriciarum OX=4758 GN=XYNA PE=1 SV=1 |
| P51584 | 2.62e-103 | 275 | 1091 | 32 | 742 | Endo-1,4-beta-xylanase Y OS=Acetivibrio thermocellus OX=1515 GN=xynY PE=1 SV=1 |
| Q53317 | 2.79e-78 | 17 | 426 | 18 | 407 | Xylanase/beta-glucanase OS=Ruminococcus flavefaciens OX=1265 GN=xynD PE=3 SV=2 |
| Q60037 | 3.87e-72 | 280 | 780 | 207 | 690 | Endo-1,4-beta-xylanase A OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=xynA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
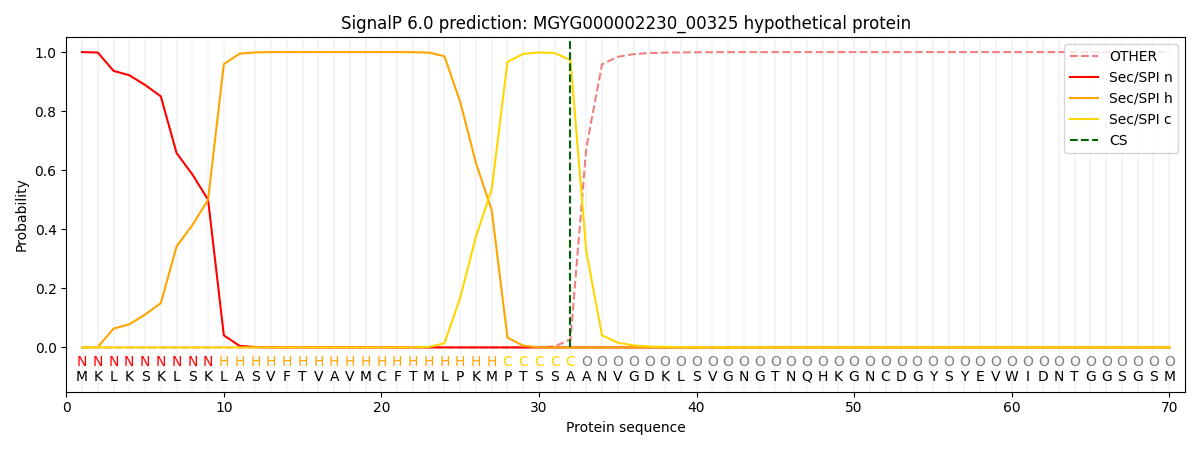
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000267 | 0.999132 | 0.000187 | 0.000162 | 0.000133 | 0.000126 |
