You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002230_00395
You are here: Home > Sequence: MGYG000002230_00395
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ruminococcus sp900545125 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus; Ruminococcus sp900545125 | |||||||||||
| CAZyme ID | MGYG000002230_00395 | |||||||||||
| CAZy Family | CBM79 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1303; End: 3384 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 394 | 662 | 2.1e-99 | 0.9891304347826086 |
| CBM79 | 94 | 204 | 3.7e-29 | 0.9545454545454546 |
| CBM79 | 231 | 331 | 7.1e-22 | 0.9545454545454546 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.74e-67 | 382 | 665 | 4 | 272 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 9.46e-36 | 365 | 669 | 45 | 367 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam18522 | DUF5620 | 8.05e-28 | 93 | 205 | 1 | 118 | Domain of unknown function (DUF5620). This is a domain of unknown function predicted to be a carbohydrate binding module. |
| pfam18522 | DUF5620 | 9.77e-17 | 230 | 332 | 1 | 118 | Domain of unknown function (DUF5620). This is a domain of unknown function predicted to be a carbohydrate binding module. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| EWM54675.1 | 4.79e-291 | 1 | 693 | 1 | 683 |
| CCZ83818.1 | 1.64e-198 | 3 | 690 | 7 | 714 |
| CDE11886.1 | 1.79e-198 | 69 | 693 | 63 | 710 |
| CDE32105.1 | 3.13e-197 | 96 | 692 | 89 | 701 |
| ERJ92483.1 | 6.24e-197 | 69 | 693 | 86 | 734 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q1I_A | 4.47e-92 | 361 | 693 | 11 | 354 | GH5-4broad specificity endoglucanase from Clostrdium longisporum [Clostridium longisporum],6Q1I_B GH5-4 broad specificity endoglucanase from Clostrdium longisporum [Clostridium longisporum] |
| 6MQ4_A | 5.54e-92 | 363 | 692 | 11 | 349 | ChainA, cellulase [Acetivibrio cellulolyticus] |
| 6PZ7_A | 1.02e-89 | 358 | 680 | 3 | 324 | GH5-4broad specificity endoglucanase from Clostridium acetobutylicum [Clostridium acetobutylicum ATCC 824] |
| 4W85_A | 3.20e-89 | 360 | 692 | 1 | 339 | Crystalstructure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in complex with glucose [uncultured bacterium],4W85_B Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in complex with glucose [uncultured bacterium],4W87_A Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from metagenomic library, in complex with a xyloglucan oligosaccharide [uncultured bacterium],4W87_B Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from metagenomic library, in complex with a xyloglucan oligosaccharide [uncultured bacterium],4W89_A Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from metagenomic library, in complex with cellotriose [uncultured bacterium],4W89_B Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from metagenomic library, in complex with cellotriose [uncultured bacterium] |
| 4W86_A | 3.73e-87 | 360 | 692 | 1 | 339 | Crystalstructure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in complex with glucose and TRIS [uncultured bacterium],4W86_B Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in complex with glucose and TRIS [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P54937 | 3.66e-90 | 361 | 693 | 36 | 379 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
| P28623 | 8.62e-86 | 364 | 691 | 43 | 371 | Endoglucanase D OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engD PE=1 SV=2 |
| P23660 | 1.85e-84 | 356 | 693 | 17 | 364 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
| P28621 | 1.88e-82 | 363 | 692 | 41 | 374 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
| Q12647 | 8.43e-77 | 363 | 679 | 24 | 349 | Endoglucanase B OS=Neocallimastix patriciarum OX=4758 GN=CELB PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
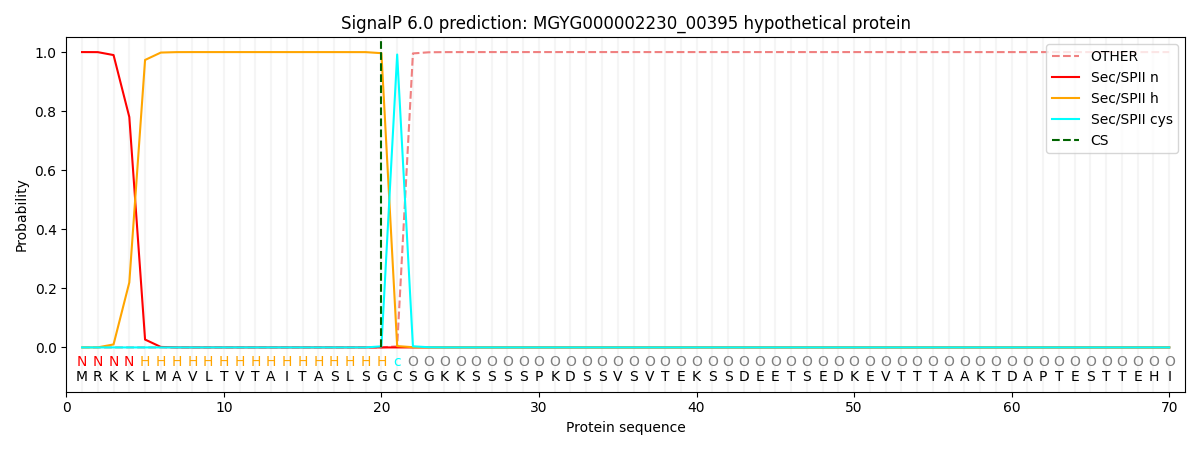
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000061 | 0.000000 | 0.000000 | 0.000000 |
