You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002247_01926
You are here: Home > Sequence: MGYG000002247_01926
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
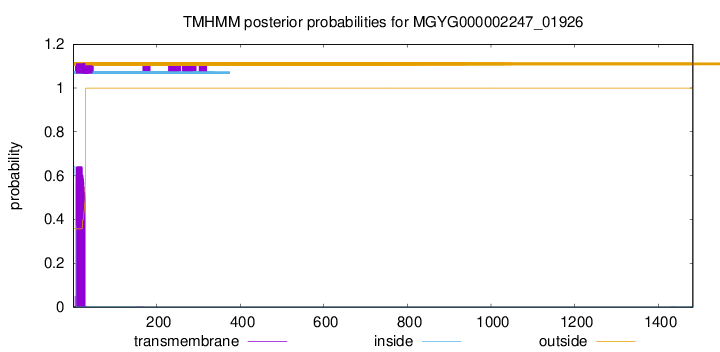
TMHMM annotations
Basic Information help
| Species | Acetatifactor sp900771995 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Acetatifactor; Acetatifactor sp900771995 | |||||||||||
| CAZyme ID | MGYG000002247_01926 | |||||||||||
| CAZy Family | CBM27 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 32766; End: 37217 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 553 | 886 | 4.6e-58 | 0.900990099009901 |
| CBM23 | 1047 | 1208 | 7.9e-43 | 0.9753086419753086 |
| CBM27 | 371 | 518 | 1.4e-25 | 0.9583333333333334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 1.37e-39 | 554 | 810 | 2 | 243 | Glycosyl hydrolase family 26. |
| COG4124 | ManB2 | 4.16e-18 | 705 | 896 | 169 | 352 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| cd00118 | LysM | 4.73e-11 | 1438 | 1482 | 2 | 45 | Lysin Motif is a small domain involved in binding peptidoglycan. LysM, a small globular domain with approximately 40 amino acids, is a widespread protein module involved in binding peptidoglycan in bacteria and chitin in eukaryotes. The domain was originally identified in enzymes that degrade bacterial cell walls, but proteins involved in many other biological functions also contain this domain. It has been reported that the LysM domain functions as a signal for specific plant-bacteria recognition in bacterial pathogenesis. Many of these enzymes are modular and are composed of catalytic units linked to one or several repeats of LysM domains. LysM domains are found in bacteria and eukaryotes. |
| smart00257 | LysM | 6.92e-10 | 1439 | 1482 | 2 | 44 | Lysin motif. |
| pfam01476 | LysM | 8.40e-09 | 1439 | 1482 | 1 | 42 | LysM domain. The LysM (lysin motif) domain is about 40 residues long. It is found in a variety of enzymes involved in bacterial cell wall degradation. This domain may have a general peptidoglycan binding function. The structure of this domain is known. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK83362.1 | 0.0 | 85 | 1247 | 265 | 1453 |
| EEV00397.1 | 0.0 | 270 | 1240 | 116 | 1086 |
| VCV20359.1 | 0.0 | 270 | 1240 | 104 | 1074 |
| CBL14297.1 | 0.0 | 270 | 1240 | 104 | 1074 |
| CBL07467.1 | 0.0 | 270 | 1240 | 104 | 1074 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4YN5_A | 6.46e-53 | 551 | 936 | 50 | 431 | Catalyticdomain of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase [Bacillus sp. JAMB750] |
| 1R7O_A | 1.60e-48 | 532 | 887 | 3 | 334 | CrystalStructure of apo-mannanase 26A from Psudomonas cellulosa [Cellvibrio japonicus] |
| 1J9Y_A | 4.23e-48 | 547 | 887 | 6 | 324 | Crystalstructure of mannanase 26A from Pseudomonas cellulosa [Cellvibrio japonicus] |
| 2WHM_A | 1.94e-47 | 547 | 887 | 6 | 324 | Cellvibriojaponicus Man26A E121A and E320G double mutant in complex with mannobiose [Cellvibrio japonicus] |
| 1GW1_A | 2.11e-47 | 547 | 887 | 2 | 320 | Substratedistortion by beta-mannanase from Pseudomonas cellulosa [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A1A278 | 1.41e-48 | 550 | 1208 | 40 | 696 | Mannan endo-1,4-beta-mannosidase OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=BAD_1030 PE=1 SV=1 |
| P49424 | 1.70e-47 | 532 | 887 | 31 | 362 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=manA PE=1 SV=2 |
| Q5AWB7 | 4.85e-16 | 608 | 851 | 76 | 313 | Probable mannan endo-1,4-beta-mannosidase E OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manE PE=3 SV=1 |
| P49425 | 5.79e-15 | 508 | 800 | 105 | 379 | Mannan endo-1,4-beta-mannosidase OS=Rhodothermus marinus (strain ATCC 43812 / DSM 4252 / R-10) OX=518766 GN=manA PE=1 SV=3 |
| P16699 | 3.49e-14 | 552 | 798 | 34 | 270 | Mannan endo-1,4-beta-mannosidase A and B OS=Caldalkalibacillus mannanilyticus (strain DSM 16130 / CIP 109019 / JCM 10596 / AM-001) OX=1236954 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
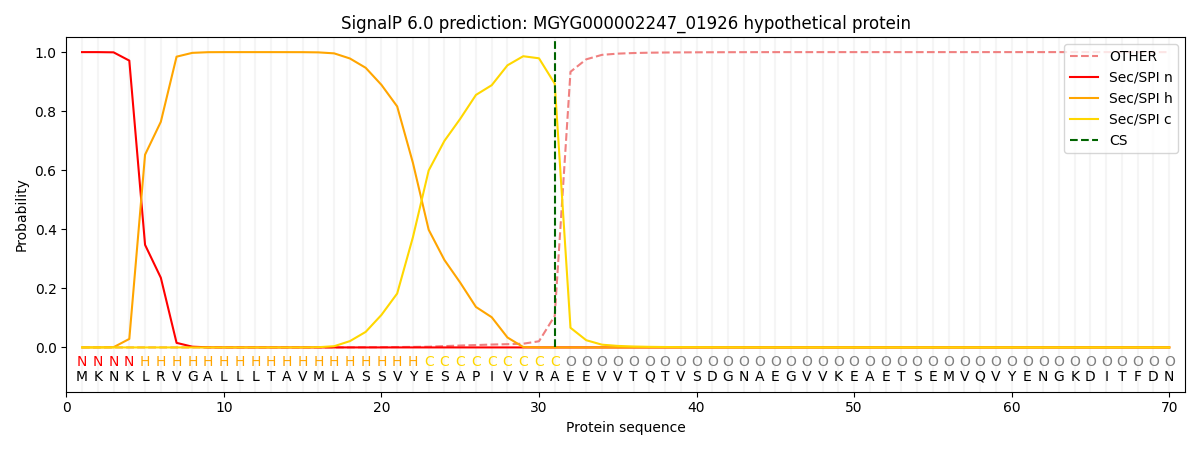
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000295 | 0.999003 | 0.000191 | 0.000189 | 0.000151 | 0.000136 |