You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002270_04183
You are here: Home > Sequence: MGYG000002270_04183
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Enterococcus_A avium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Enterococcaceae; Enterococcus_A; Enterococcus_A avium | |||||||||||
| CAZyme ID | MGYG000002270_04183 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2312; End: 6970 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5412 | COG5412 | 2.46e-09 | 740 | 1241 | 1 | 493 | Phage-related protein [Mobilome: prophages, transposons]. |
| COG5412 | COG5412 | 2.34e-07 | 574 | 1005 | 102 | 494 | Phage-related protein [Mobilome: prophages, transposons]. |
| pfam01442 | Apolipoprotein | 8.80e-07 | 1030 | 1190 | 1 | 165 | Apolipoprotein A1/A4/E domain. These proteins contain several 22 residue repeats which form a pair of alpha helices. This family includes: Apolipoprotein A-I. Apolipoprotein A-IV. Apolipoprotein E. |
| TIGR02168 | SMC_prok_B | 1.49e-04 | 3 | 308 | 188 | 503 | chromosome segregation protein SMC, common bacterial type. SMC (structural maintenance of chromosomes) proteins bind DNA and act in organizing and segregating chromosomes for partition. SMC proteins are found in bacteria, archaea, and eukaryotes. This family represents the SMC protein of most bacteria. The smc gene is often associated with scpB (TIGR00281) and scpA genes, where scp stands for segregation and condensation protein. SMC was shown (in Caulobacter crescentus) to be induced early in S phase but present and bound to DNA throughout the cell cycle. [Cellular processes, Cell division, DNA metabolism, Chromosome-associated proteins] |
| COG5185 | HEC1 | 7.23e-04 | 161 | 331 | 307 | 463 | Protein involved in chromosome segregation, interacts with SMC proteins [Cell cycle control, cell division, chromosome partitioning]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCQ10945.1 | 0.0 | 1 | 1552 | 1 | 1546 |
| AXG89996.1 | 1.30e-204 | 1 | 1375 | 1 | 1245 |
| ACZ64075.1 | 1.90e-204 | 1 | 1375 | 3 | 1247 |
| ACZ64141.1 | 1.90e-204 | 1 | 1375 | 3 | 1247 |
| QKR90302.1 | 7.21e-204 | 1 | 1375 | 3 | 1247 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000065 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
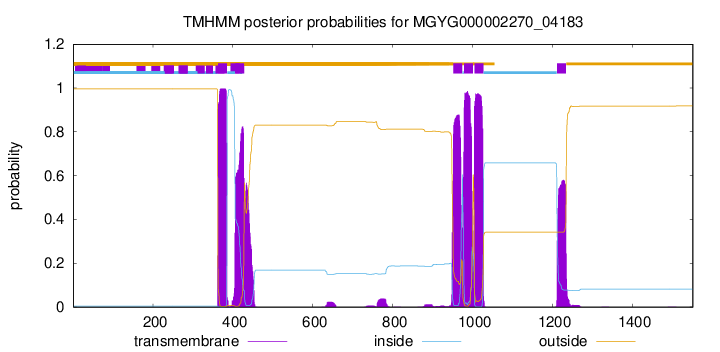
| start | end |
|---|---|
| 363 | 385 |
| 406 | 428 |
| 952 | 974 |
| 979 | 1001 |
| 1005 | 1027 |
| 1212 | 1234 |
