You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002273_00766
You are here: Home > Sequence: MGYG000002273_00766
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides stercorirosoris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides stercorirosoris | |||||||||||
| CAZyme ID | MGYG000002273_00766 | |||||||||||
| CAZy Family | PL8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 136904; End: 137929 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam09092 | Lyase_N | 3.97e-53 | 20 | 179 | 2 | 166 | Lyase, N terminal. Members of this family are predominantly found in chondroitin ABC lyase I, and adopt a jelly-roll fold topology consisting of a two-layered bent beta-sheet sandwich with one short alpha-helix. The convex beta sheet is composed of five antiparallel strands, whilst the concave beta-sheet contains five antiparallel beta-strands with a loop between two consecutive strands folding back onto the concave surface. This domain is required for binding of the protein to long glycosaminoglycan chains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDO68088.1 | 2.39e-218 | 1 | 309 | 1 | 309 |
| ALJ59760.1 | 2.59e-204 | 1 | 309 | 1 | 309 |
| QUT89208.1 | 5.45e-198 | 1 | 309 | 1 | 309 |
| QRQ49161.1 | 1.18e-131 | 1 | 309 | 1 | 306 |
| QUT44903.1 | 1.28e-130 | 1 | 309 | 1 | 306 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2Q1F_A | 6.12e-23 | 35 | 250 | 35 | 252 | Crystalstructure of chondroitin sulfate lyase abc from bacteroides thetaiotaomicron wal2926 [Bacteroides thetaiotaomicron],2Q1F_B Crystal structure of chondroitin sulfate lyase abc from bacteroides thetaiotaomicron wal2926 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8A2I1 | 5.58e-23 | 35 | 250 | 35 | 252 | Chondroitin sulfate ABC exolyase OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=chonabc PE=1 SV=1 |
| C5G6D7 | 7.52e-23 | 35 | 250 | 35 | 252 | Chondroitin sulfate ABC exolyase OS=Bacteroides thetaiotaomicron OX=818 GN=chonabc PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
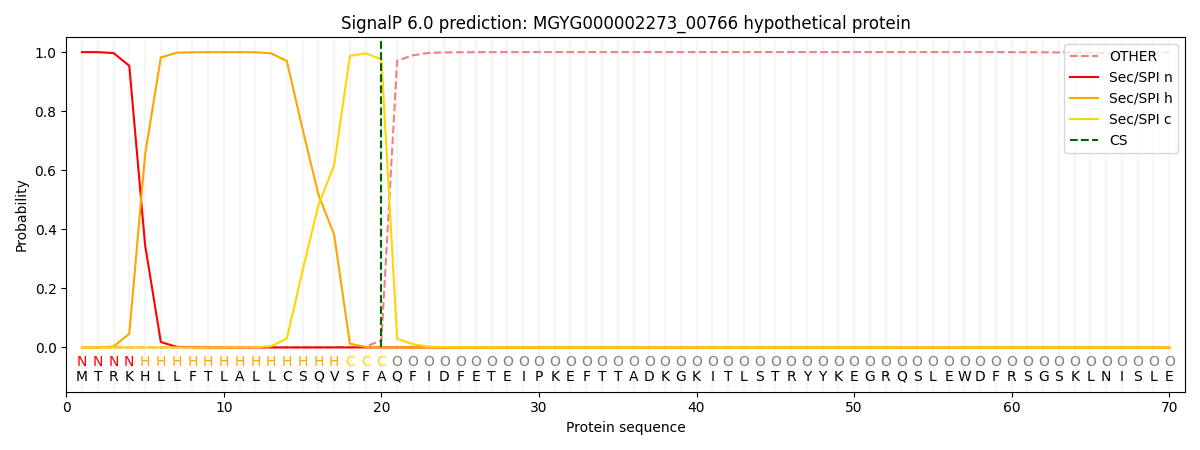
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002807 | 0.994523 | 0.000613 | 0.000783 | 0.000600 | 0.000627 |
