You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002273_02408
You are here: Home > Sequence: MGYG000002273_02408
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides stercorirosoris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides stercorirosoris | |||||||||||
| CAZyme ID | MGYG000002273_02408 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 122899; End: 124431 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 206 | 487 | 3.2e-43 | 0.9583333333333334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02304 | PLN02304 | 2.53e-13 | 193 | 499 | 66 | 368 | probable pectinesterase |
| PLN02480 | PLN02480 | 7.89e-12 | 202 | 505 | 46 | 340 | Probable pectinesterase |
| PLN02682 | PLN02682 | 2.56e-11 | 205 | 481 | 70 | 347 | pectinesterase family protein |
| PLN02671 | PLN02671 | 8.37e-11 | 206 | 481 | 61 | 337 | pectinesterase |
| PLN02773 | PLN02773 | 1.20e-10 | 206 | 496 | 7 | 298 | pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ61700.1 | 0.0 | 2 | 510 | 1 | 509 |
| QUT92737.1 | 0.0 | 2 | 510 | 1 | 509 |
| AII65147.1 | 6.88e-306 | 8 | 507 | 1 | 500 |
| QJR79100.1 | 6.88e-306 | 8 | 507 | 1 | 500 |
| QJR66174.1 | 6.88e-306 | 8 | 507 | 1 | 500 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O23038 | 6.38e-12 | 196 | 509 | 78 | 393 | Probable pectinesterase 8 OS=Arabidopsis thaliana OX=3702 GN=PME8 PE=2 SV=2 |
| Q9ZQA3 | 1.19e-10 | 191 | 369 | 77 | 250 | Probable pectinesterase 15 OS=Arabidopsis thaliana OX=3702 GN=PME15 PE=2 SV=1 |
| Q5MFV8 | 2.74e-08 | 195 | 459 | 273 | 545 | Pectinesterase 5 OS=Arabidopsis thaliana OX=3702 GN=PME5 PE=1 SV=2 |
| Q9FM79 | 7.42e-08 | 185 | 446 | 58 | 323 | Pectinesterase QRT1 OS=Arabidopsis thaliana OX=3702 GN=QRT1 PE=1 SV=1 |
| Q3E9D3 | 3.33e-07 | 206 | 483 | 29 | 305 | Probable pectinesterase 55 OS=Arabidopsis thaliana OX=3702 GN=PME55 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
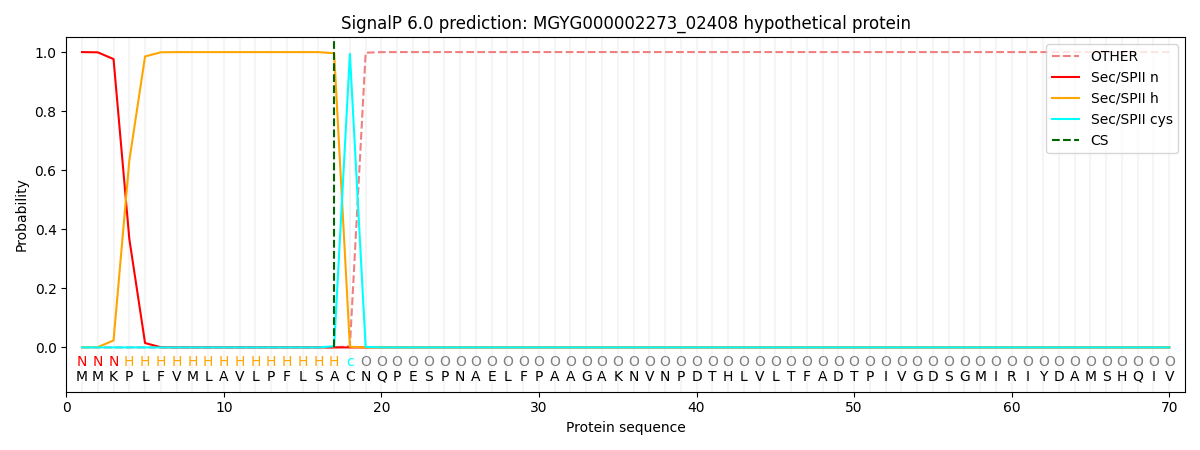
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000073 | 0.000000 | 0.000000 | 0.000000 |
