You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002281_01341
You are here: Home > Sequence: MGYG000002281_01341
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides faecis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides faecis | |||||||||||
| CAZyme ID | MGYG000002281_01341 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 242403; End: 243980 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 26 | 519 | 8.7e-198 | 0.9917355371900827 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14587 | Glyco_hydr_30_2 | 8.35e-149 | 28 | 388 | 6 | 366 | O-Glycosyl hydrolase family 30. |
| COG5520 | XynC | 2.02e-12 | 28 | 480 | 38 | 411 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam17189 | Glyco_hydro_30C | 2.36e-06 | 420 | 519 | 1 | 63 | Glycosyl hydrolase family 30 beta sandwich domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCA52365.1 | 0.0 | 1 | 521 | 5 | 525 |
| QUT73144.1 | 0.0 | 1 | 524 | 5 | 528 |
| QUT40997.1 | 0.0 | 1 | 524 | 5 | 528 |
| QMW85467.1 | 0.0 | 5 | 524 | 9 | 528 |
| AAO78149.1 | 0.0 | 5 | 524 | 9 | 528 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CLW_A | 1.06e-89 | 28 | 520 | 10 | 495 | Crystalstructure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_B Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_C Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_D Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_E Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_F Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 6M5Z_A | 9.50e-15 | 35 | 520 | 34 | 452 | ChainA, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612],6M5Z_B Chain B, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612] |
| 7O0E_A | 1.22e-13 | 28 | 523 | 7 | 449 | ChainA, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464],7O0E_G Chain G, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464] |
| 7NCX_AAA | 7.35e-13 | 28 | 523 | 14 | 456 | ChainAAA, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464] |
| 1NOF_A | 4.62e-09 | 141 | 522 | 77 | 383 | ChainA, xylanase [Dickeya chrysanthemi],2Y24_A Chain A, XYLANASE [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q76FP5 | 7.75e-15 | 36 | 263 | 34 | 268 | Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1 |
| G2Q1N4 | 1.32e-13 | 28 | 523 | 31 | 473 | GH30 family xylanase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Xyn30A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
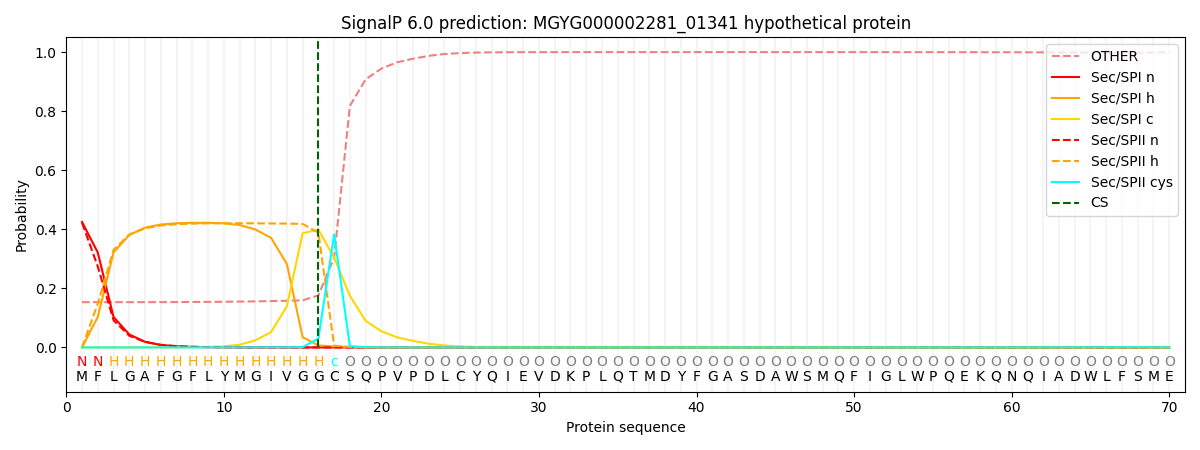
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.166950 | 0.405853 | 0.426227 | 0.000290 | 0.000296 | 0.000393 |
