You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002281_02402
You are here: Home > Sequence: MGYG000002281_02402
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides faecis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides faecis | |||||||||||
| CAZyme ID | MGYG000002281_02402 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11607; End: 14717 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 51 | 911 | 5.2e-221 | 0.9773936170212766 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09525 | lacZ | 0.0 | 25 | 961 | 15 | 964 | beta-galactosidase. |
| PRK10340 | ebgA | 0.0 | 25 | 1031 | 2 | 1000 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| COG3250 | LacZ | 2.48e-166 | 66 | 913 | 14 | 808 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| smart01038 | Bgal_small_N | 2.61e-111 | 756 | 1028 | 1 | 272 | Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase. |
| pfam02836 | Glyco_hydro_2_C | 1.75e-97 | 339 | 620 | 1 | 300 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMW85735.1 | 0.0 | 1 | 1036 | 1 | 1036 |
| AAO78446.1 | 0.0 | 1 | 1036 | 1 | 1036 |
| QQA08535.1 | 0.0 | 1 | 1036 | 1 | 1036 |
| ALJ44474.1 | 0.0 | 1 | 1036 | 1 | 1036 |
| QUT41248.1 | 0.0 | 1 | 1036 | 1 | 1036 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 5.72e-241 | 26 | 1029 | 3 | 980 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 5.90e-241 | 26 | 1029 | 4 | 981 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 3BGA_A | 5.10e-218 | 22 | 1031 | 5 | 1001 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 3DEC_A | 6.53e-216 | 26 | 1031 | 6 | 997 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 1JZ7_A | 6.34e-159 | 26 | 1027 | 15 | 1019 | E.COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_B E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_C E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_D E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],4TTG_A Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_B Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_C Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_D Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 3.23e-240 | 26 | 1029 | 4 | 981 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| O52847 | 1.18e-219 | 21 | 1031 | 14 | 1034 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| Q9K9C6 | 6.69e-195 | 23 | 1031 | 6 | 1014 | Beta-galactosidase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=lacZ PE=3 SV=1 |
| Q6LL68 | 1.16e-185 | 26 | 1030 | 11 | 1027 | Beta-galactosidase OS=Photobacterium profundum (strain SS9) OX=298386 GN=lacZ PE=3 SV=1 |
| P23989 | 8.71e-184 | 29 | 1031 | 13 | 1025 | Beta-galactosidase OS=Streptococcus thermophilus OX=1308 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
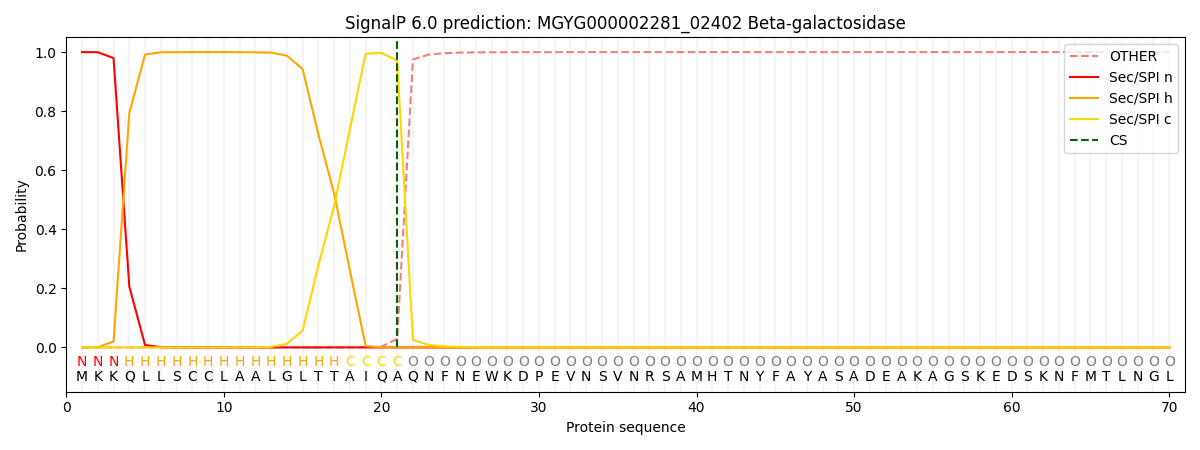
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000353 | 0.998823 | 0.000221 | 0.000211 | 0.000191 | 0.000175 |
