You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002281_03242
You are here: Home > Sequence: MGYG000002281_03242
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides faecis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides faecis | |||||||||||
| CAZyme ID | MGYG000002281_03242 | |||||||||||
| CAZy Family | GH141 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 83296; End: 85632 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH141 | 21 | 535 | 1e-58 | 0.9886148007590133 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 3.27e-08 | 306 | 452 | 23 | 155 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 3.33e-06 | 307 | 450 | 1 | 129 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| cd00987 | PDZ_serine_protease | 7.12e-05 | 699 | 775 | 14 | 90 | PDZ domain of trypsin-like serine proteases, such as DegP/HtrA, which are oligomeric proteins involved in heat-shock response, chaperone function, and apoptosis. May be responsible for substrate recognition and/or binding, as most PDZ domains bind C-terminal polypeptides, though binding to internal (non-C-terminal) polypeptides and even to lipids has been demonstrated. In this subfamily of protease-associated PDZ domains a C-terminal beta-strand forms the peptide-binding groove base, a circular permutation with respect to PDZ domains found in Eumetazoan signaling proteins. |
| pfam13229 | Beta_helix | 0.006 | 272 | 365 | 64 | 151 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWW32784.1 | 7.69e-199 | 6 | 745 | 16 | 767 |
| BAU63437.1 | 1.52e-44 | 22 | 556 | 57 | 593 |
| AFZ35556.1 | 1.19e-42 | 56 | 655 | 97 | 710 |
| QZT37819.1 | 4.07e-33 | 2 | 646 | 3 | 708 |
| BBD44453.1 | 4.90e-33 | 1 | 664 | 1 | 666 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQP_A | 3.01e-17 | 23 | 444 | 28 | 501 | Glycosidehydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_B Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_C Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_D Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_E Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_F Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_G Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_H Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
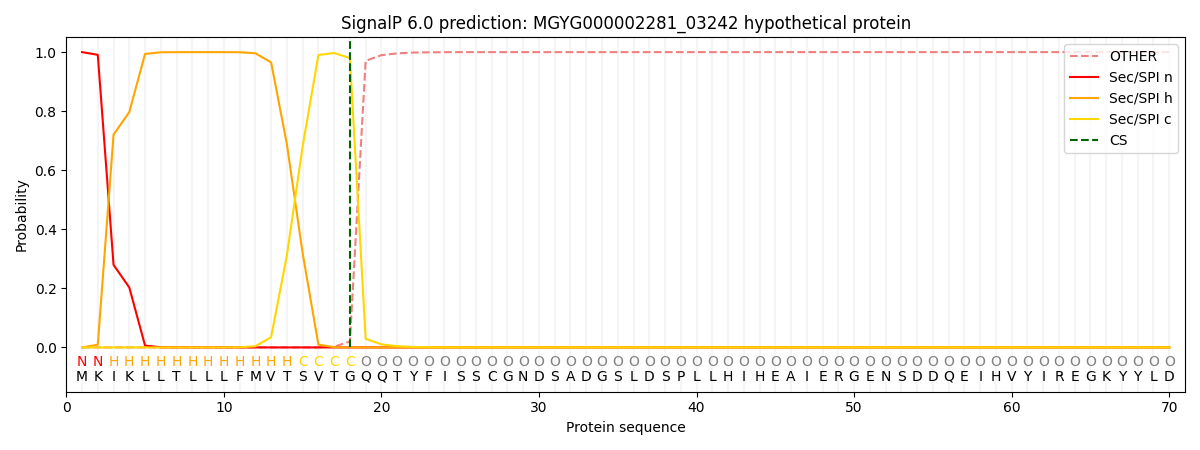
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000360 | 0.998956 | 0.000214 | 0.000156 | 0.000147 | 0.000148 |
