You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002281_03516
You are here: Home > Sequence: MGYG000002281_03516
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides faecis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides faecis | |||||||||||
| CAZyme ID | MGYG000002281_03516 | |||||||||||
| CAZy Family | GH143 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 54908; End: 58237 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH143 | 21 | 570 | 7.4e-292 | 0.9946043165467626 |
| GH142 | 633 | 1101 | 2e-231 | 0.9979123173277662 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13088 | BNR_2 | 1.31e-08 | 269 | 343 | 168 | 247 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| COG3408 | GDB1 | 8.01e-06 | 754 | 1010 | 343 | 589 | Glycogen debranching enzyme (alpha-1,6-glucosidase) [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMW88040.1 | 0.0 | 1 | 1109 | 1 | 1109 |
| QQA06618.1 | 0.0 | 1 | 1109 | 1 | 1109 |
| AAO76127.1 | 0.0 | 1 | 1109 | 1 | 1109 |
| ALJ42177.1 | 0.0 | 1 | 1109 | 1 | 1109 |
| QUT38404.1 | 0.0 | 1 | 1109 | 1 | 1109 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQR_A | 0.0 | 21 | 1109 | 20 | 1108 | SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
| 5MQS_A | 0.0 | 21 | 1109 | 20 | 1108 | SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
| 6M5A_A | 1.11e-17 | 652 | 1107 | 303 | 814 | Crystalstructure of GH121 beta-L-arabinobiosidase HypBA2 from Bifidobacterium longum [Bifidobacterium longum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH9 | 8.72e-17 | 652 | 1107 | 334 | 845 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
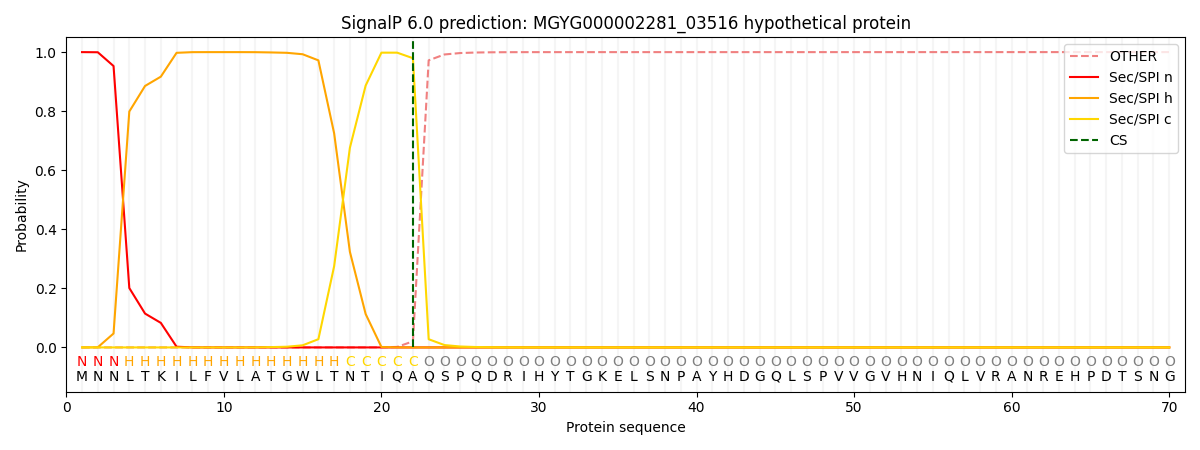
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000200 | 0.999183 | 0.000155 | 0.000153 | 0.000139 | 0.000125 |
