You are browsing environment: HUMAN GUT
MGYG000002281_03530
Basic Information
help
Species
Bacteroides faecis
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides faecis
CAZyme ID
MGYG000002281_03530
CAZy Family
GH163
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000002281
6145961
Isolate
China
Asia
Gene Location
Start: 83191;
End: 85395
Strand: -
No EC number prediction in MGYG000002281_03530.
Family
Start
End
Evalue
family coverage
GH163
197
451
1.5e-110
0.9960159362549801
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam16126
DUF4838
2.34e-131
190
451
1
263
Domain of unknown function (DUF4838). This family consists of several uncharacterized proteins found in various Bacteroides and Chloroflexus species. The function of this family is unknown.
more
pfam00754
F5_F8_type_C
4.74e-09
599
726
13
125
F5/8 type C domain. This domain is also known as the discoidin (DS) domain family.
more
pfam02838
Glyco_hydro_20b
3.40e-05
21
121
5
105
Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
6Q63_A
7.71e-14
590
734
629
774
BT0459[Bacteroides thetaiotaomicron],6Q63_B BT0459 [Bacteroides thetaiotaomicron],6Q63_C BT0459 [Bacteroides thetaiotaomicron]
more
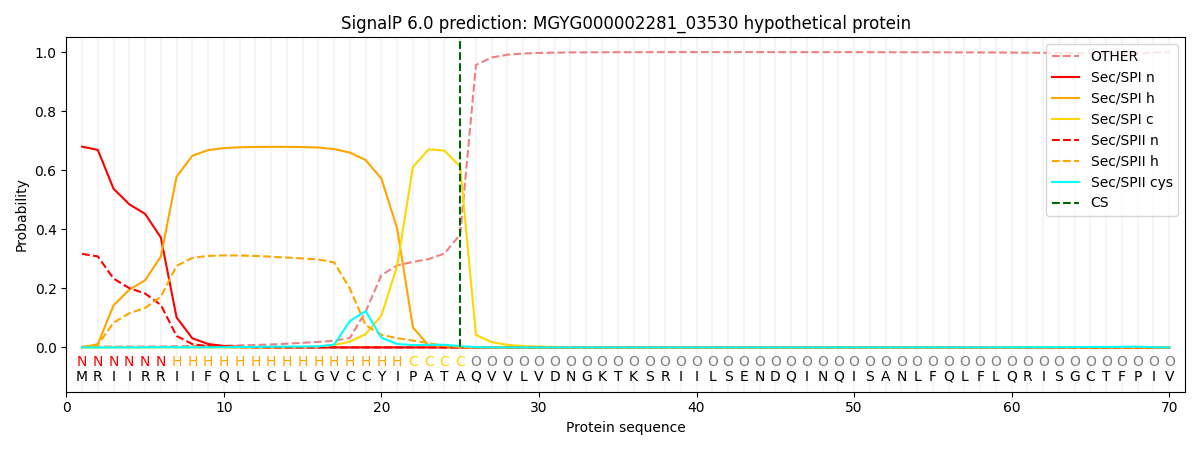
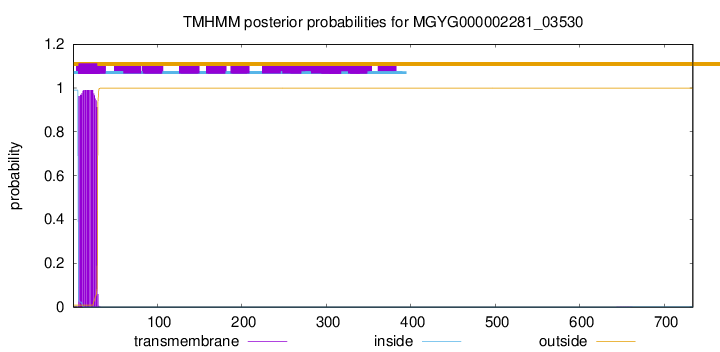
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.005033
0.668319
0.324927
0.000991
0.000364
0.000326