You are browsing environment: HUMAN GUT
MGYG000002282_03437
Basic Information
help
Species
Parabacteroides gordonii
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides gordonii
CAZyme ID
MGYG000002282_03437
CAZy Family
GH163
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
730
82505.06
5.706
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000002282
6348794
Isolate
China
Asia
Gene Location
Start: 131214;
End: 133406
Strand: +
No EC number prediction in MGYG000002282_03437.
Family
Start
End
Evalue
family coverage
GH163
194
446
1.8e-105
0.9880478087649402
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam16126
DUF4838
1.02e-135
186
446
1
262
Domain of unknown function (DUF4838). This family consists of several uncharacterized proteins found in various Bacteroides and Chloroflexus species. The function of this family is unknown.
more
pfam02838
Glyco_hydro_20b
9.82e-08
18
117
20
105
Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold.
more
pfam00754
F5_F8_type_C
6.93e-07
612
722
25
125
F5/8 type C domain. This domain is also known as the discoidin (DS) domain family.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
6Q63_A
1.08e-11
580
730
622
774
BT0459[Bacteroides thetaiotaomicron],6Q63_B BT0459 [Bacteroides thetaiotaomicron],6Q63_C BT0459 [Bacteroides thetaiotaomicron]
more
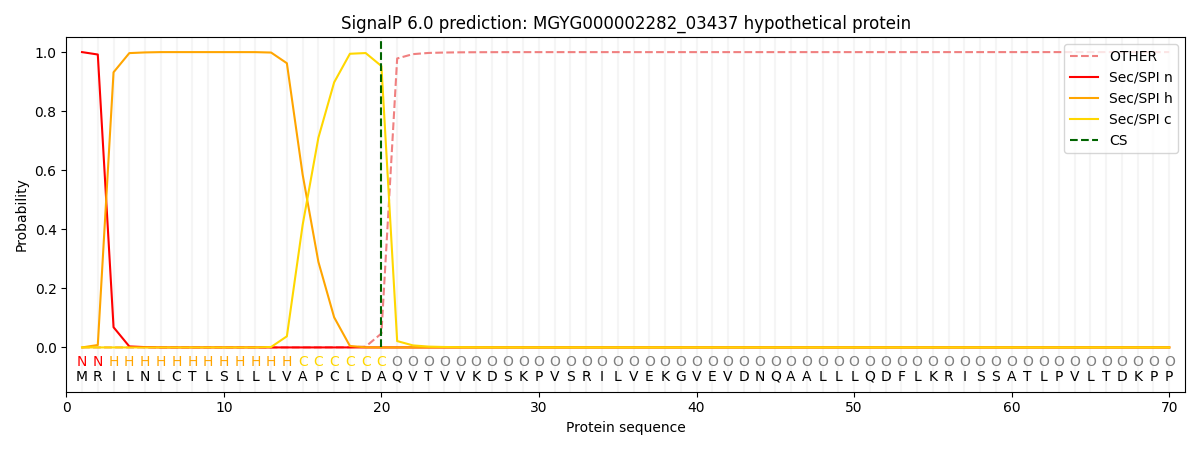
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000227
0.999128
0.000167
0.000147
0.000143
0.000141
There is no transmembrane helices in MGYG000002282_03437.