You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002282_03594
You are here: Home > Sequence: MGYG000002282_03594
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides gordonii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides gordonii | |||||||||||
| CAZyme ID | MGYG000002282_03594 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 64302; End: 65399 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 35 | 348 | 1.6e-88 | 0.8519553072625698 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03663 | Glyco_hydro_76 | 1.84e-49 | 51 | 299 | 35 | 290 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| COG4833 | COG4833 | 7.04e-23 | 104 | 320 | 88 | 306 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
| COG1331 | YyaL | 3.99e-05 | 110 | 327 | 274 | 507 | Uncharacterized conserved protein YyaL, SSP411 family, contains thoiredoxin and six-hairpin glycosidase-like domains [General function prediction only]. |
| COG4225 | YesR | 2.24e-04 | 224 | 323 | 31 | 114 | Rhamnogalacturonyl hydrolase YesR [Carbohydrate transport and metabolism]. |
| cd04791 | LanC_SerThrkinase | 2.78e-04 | 115 | 261 | 92 | 234 | Lanthionine synthetase C-like domain associated with serine/threonine kinases. Some members of this subgroup lack the zinc binding site and the active site residues, and therefore are most likely inactive. The function of this domain is unknown. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIX66371.1 | 3.86e-242 | 1 | 364 | 1 | 364 |
| QJE28248.1 | 2.07e-241 | 1 | 363 | 1 | 363 |
| BBK90205.1 | 4.18e-241 | 1 | 363 | 1 | 363 |
| QUR49840.1 | 1.70e-240 | 1 | 363 | 1 | 363 |
| AST52979.1 | 1.98e-239 | 1 | 363 | 1 | 363 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3K7X_A | 3.85e-34 | 104 | 321 | 87 | 299 | ChainA, Lin0763 protein [Listeria innocua] |
| 6SHD_A | 1.78e-28 | 25 | 320 | 62 | 348 | Structureof the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_B Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_C Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6] |
| 6Y8F_A | 6.47e-28 | 25 | 320 | 63 | 349 | ChainA, Alpha-1,6-endo-mannanase GH76A mutant [Salegentibacter sp. Hel_I_6] |
| 6SHM_A | 2.15e-26 | 25 | 320 | 63 | 349 | Aninactive (D136A and D137A) variant of alpha-1,6-mannanase, GH76A of Salegentibacter sp. HEL1_6 in complex with alpha-1,6-mannotetrose [Salegentibacter sp. Hel_I_6] |
| 4MU9_A | 3.27e-21 | 107 | 357 | 116 | 361 | Crystalstructure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482],4MU9_B Crystal structure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
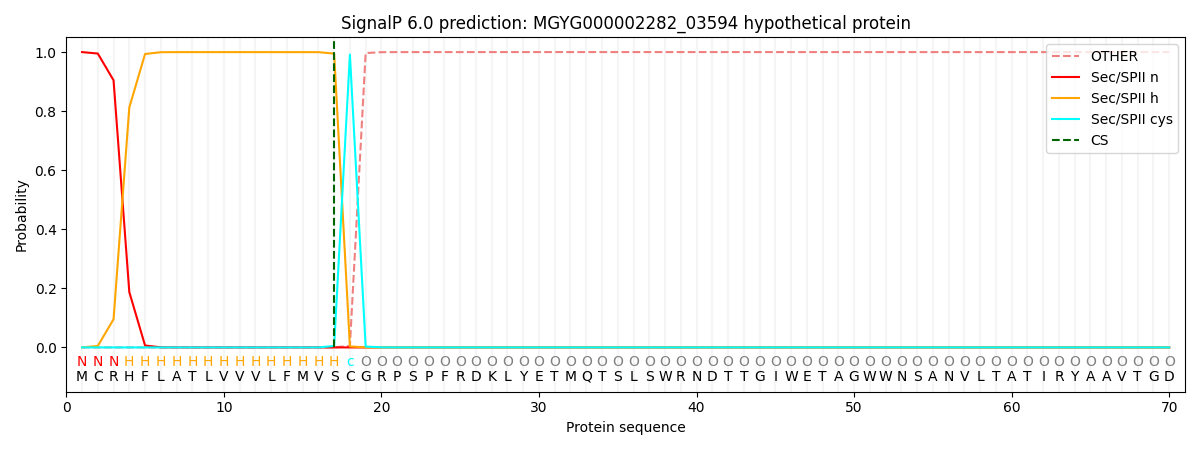
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000039 | 0.000000 | 0.000000 | 0.000000 |
