You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002282_04133
You are here: Home > Sequence: MGYG000002282_04133
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides gordonii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides gordonii | |||||||||||
| CAZyme ID | MGYG000002282_04133 | |||||||||||
| CAZy Family | GH141 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 166026; End: 168398 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH141 | 26 | 487 | 7e-58 | 0.9259962049335864 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd00987 | PDZ_serine_protease | 1.16e-07 | 695 | 775 | 2 | 81 | PDZ domain of trypsin-like serine proteases, such as DegP/HtrA, which are oligomeric proteins involved in heat-shock response, chaperone function, and apoptosis. May be responsible for substrate recognition and/or binding, as most PDZ domains bind C-terminal polypeptides, though binding to internal (non-C-terminal) polypeptides and even to lipids has been demonstrated. In this subfamily of protease-associated PDZ domains a C-terminal beta-strand forms the peptide-binding groove base, a circular permutation with respect to PDZ domains found in Eumetazoan signaling proteins. |
| pfam13229 | Beta_helix | 8.06e-06 | 422 | 584 | 1 | 156 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13180 | PDZ_2 | 8.09e-06 | 715 | 775 | 3 | 63 | PDZ domain. |
| pfam00595 | PDZ | 4.33e-05 | 715 | 773 | 22 | 80 | PDZ domain (Also known as DHR or GLGF). PDZ domains are found in diverse signaling proteins. |
| smart00228 | PDZ | 1.67e-04 | 701 | 775 | 9 | 83 | Domain present in PSD-95, Dlg, and ZO-1/2. Also called DHR (Dlg homologous region) or GLGF (relatively well conserved tetrapeptide in these domains). Some PDZs have been shown to bind C-terminal polypeptides; others appear to bind internal (non-C-terminal) polypeptides. Different PDZs possess different binding specificities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWW32784.1 | 9.25e-219 | 2 | 760 | 6 | 773 |
| BAU63437.1 | 9.92e-38 | 50 | 664 | 84 | 718 |
| AQQ71020.1 | 6.70e-35 | 22 | 666 | 19 | 711 |
| AWV97657.1 | 3.74e-34 | 27 | 663 | 29 | 707 |
| QYR21751.1 | 3.08e-33 | 17 | 469 | 398 | 850 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
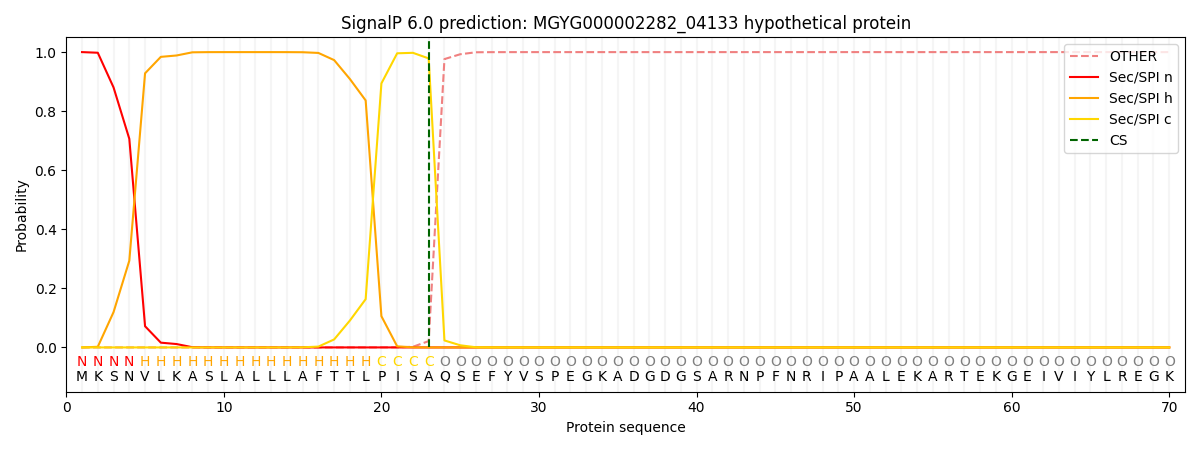
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000342 | 0.998788 | 0.000256 | 0.000200 | 0.000192 | 0.000188 |
