You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002291_01048
You are here: Home > Sequence: MGYG000002291_01048
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | OM05-12 sp003438995 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; OM05-12; OM05-12 sp003438995 | |||||||||||
| CAZyme ID | MGYG000002291_01048 | |||||||||||
| CAZy Family | PL38 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 76130; End: 77203 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL38 | 145 | 293 | 5.4e-18 | 0.5483870967741935 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05426 | Alginate_lyase | 9.67e-42 | 79 | 293 | 44 | 274 | Alginate lyase. This family contains several bacterial alginate lyase proteins. Alginate is a family of 1-4-linked copolymers of beta -D-mannuronic acid (M) and alpha -L-guluronic acid (G). It is produced by brown algae and by some bacteria belonging to the genera Azotobacter and Pseudomonas. Alginate lyases catalyze the depolymerization of alginates by beta -elimination, generating a molecule containing 4-deoxy-L-erythro-hex-4-enepyranosyluronate at the nonreducing end. This family adopts an all alpha fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT50612.1 | 2.01e-162 | 1 | 357 | 1 | 359 |
| BAM95908.1 | 3.58e-66 | 4 | 353 | 14 | 372 |
| BAM95907.1 | 1.60e-54 | 34 | 352 | 43 | 376 |
| QCP58556.1 | 7.10e-14 | 77 | 297 | 108 | 350 |
| ARB85706.1 | 8.18e-14 | 77 | 295 | 137 | 377 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
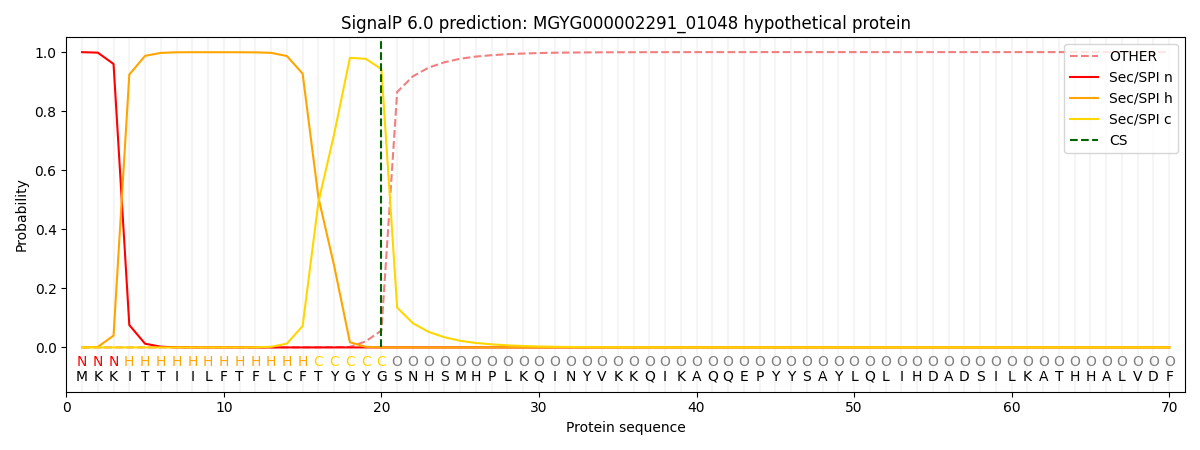
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000775 | 0.998104 | 0.000544 | 0.000191 | 0.000188 | 0.000182 |
