You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002291_02619
You are here: Home > Sequence: MGYG000002291_02619
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | OM05-12 sp003438995 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; OM05-12; OM05-12 sp003438995 | |||||||||||
| CAZyme ID | MGYG000002291_02619 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 53850; End: 57527 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 745 | 1113 | 4.2e-35 | 0.9166666666666666 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 1.82e-18 | 734 | 1108 | 80 | 389 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02217 | PLN02217 | 1.11e-12 | 836 | 1168 | 290 | 592 | probable pectinesterase/pectinesterase inhibitor |
| PRK10531 | PRK10531 | 2.44e-11 | 748 | 1104 | 95 | 410 | putative acyl-CoA thioester hydrolase. |
| PLN02170 | PLN02170 | 5.22e-11 | 860 | 1136 | 301 | 518 | probable pectinesterase/pectinesterase inhibitor |
| PLN02708 | PLN02708 | 8.09e-10 | 836 | 1136 | 282 | 550 | Probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AOW17789.1 | 0.0 | 3 | 1223 | 381 | 1604 |
| QBZ98893.1 | 0.0 | 74 | 1223 | 69 | 1217 |
| AVR45655.1 | 0.0 | 27 | 1145 | 289 | 1405 |
| AOZ99781.1 | 0.0 | 22 | 1140 | 834 | 1930 |
| APU68882.1 | 0.0 | 147 | 1145 | 514 | 1513 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3UW0_A | 1.92e-10 | 838 | 1058 | 82 | 296 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9SKX2 | 1.85e-09 | 901 | 1131 | 287 | 502 | Probable pectinesterase/pectinesterase inhibitor 16 OS=Arabidopsis thaliana OX=3702 GN=PME16 PE=2 SV=1 |
| P09607 | 9.24e-08 | 918 | 1134 | 332 | 536 | Pectinesterase 2.1 OS=Solanum lycopersicum OX=4081 GN=PME2.1 PE=2 SV=2 |
| Q84R10 | 1.16e-07 | 852 | 1137 | 276 | 510 | Probable pectinesterase/pectinesterase inhibitor 36 OS=Arabidopsis thaliana OX=3702 GN=PME36 PE=2 SV=2 |
| Q1JPL7 | 1.23e-07 | 836 | 1107 | 285 | 519 | Pectinesterase/pectinesterase inhibitor 18 OS=Arabidopsis thaliana OX=3702 GN=PME18 PE=1 SV=3 |
| Q96576 | 2.09e-07 | 918 | 1114 | 326 | 513 | Pectinesterase 3 OS=Solanum lycopersicum OX=4081 GN=PME3 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
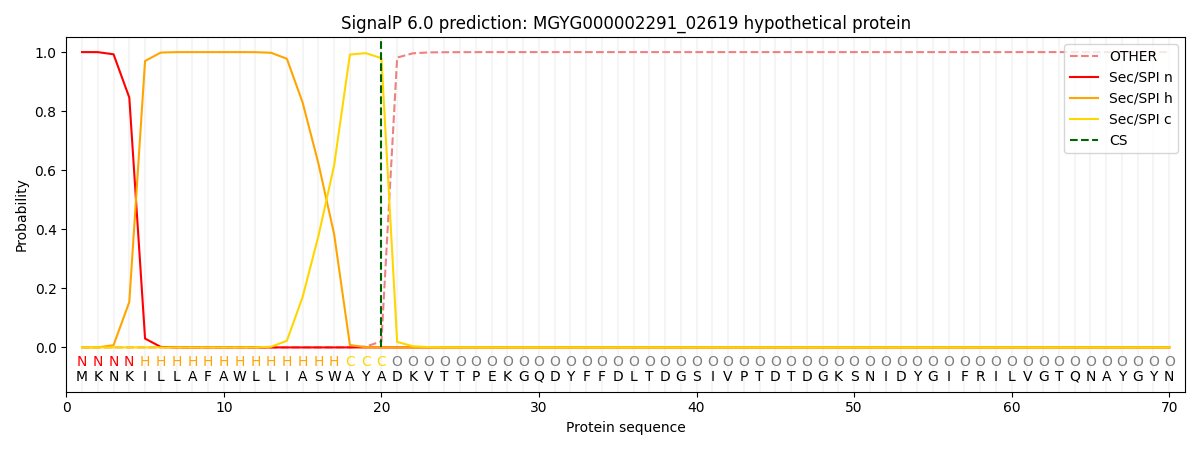
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000267 | 0.999033 | 0.000200 | 0.000168 | 0.000157 | 0.000151 |
