You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002292_02136
You are here: Home > Sequence: MGYG000002292_02136
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
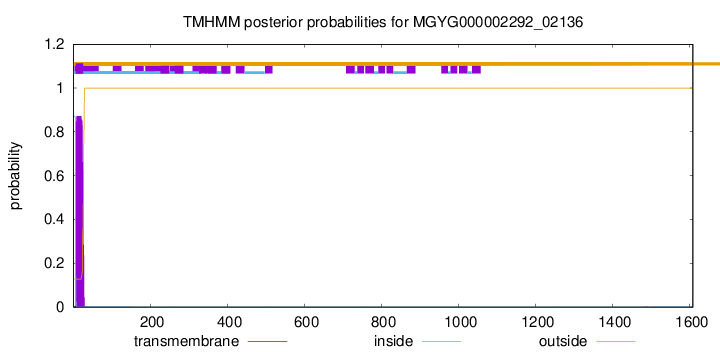
TMHMM annotations
Basic Information help
| Species | TF01-11 sp003529475 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; TF01-11; TF01-11 sp003529475 | |||||||||||
| CAZyme ID | MGYG000002292_02136 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 73875; End: 78701 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 583 | 870 | 6.4e-71 | 0.9927536231884058 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.41e-46 | 582 | 868 | 14 | 267 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.05e-17 | 547 | 744 | 33 | 232 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam04886 | PT | 2.25e-06 | 1395 | 1423 | 1 | 32 | PT repeat. This short repeat is composed on the tetrapeptide XPTX. This repeat is found in a variety of proteins, however it is not clear if these repeats are homologous to each other. The alignment represents nine copies of this repeat. |
| pfam04886 | PT | 2.82e-06 | 1392 | 1422 | 4 | 35 | PT repeat. This short repeat is composed on the tetrapeptide XPTX. This repeat is found in a variety of proteins, however it is not clear if these repeats are homologous to each other. The alignment represents nine copies of this repeat. |
| NF033186 | internalin_K | 9.90e-06 | 1307 | 1446 | 432 | 572 | class 1 internalin InlK. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface-anchored proteins with an N-terminal signal peptide, leucine-rich repeats, and a C-terminal LPXTG processing and cell surface anchoring site. Members of this family are internalin K (InlK), a virulence factor. See articles PMID:17764999. for a general discussion of internalins, and PMID:21829365, PMID:22082958, and PMID:23958637 for more information about internalin K. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUO18792.1 | 2.13e-188 | 463 | 1374 | 162 | 1050 |
| AUO19859.1 | 1.16e-114 | 462 | 1106 | 19 | 642 |
| BCN29385.1 | 1.32e-52 | 545 | 1006 | 339 | 786 |
| QNU65955.1 | 1.50e-50 | 542 | 897 | 28 | 409 |
| BAE44526.1 | 1.04e-49 | 542 | 906 | 27 | 403 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2JEP_A | 2.74e-47 | 557 | 906 | 34 | 393 | Nativefamily 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEP_B Native family 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEQ_A Family 5 xyloglucanase from Paenibacillus pabuli in complex with ligand [Paenibacillus pabuli] |
| 4IM4_A | 3.27e-46 | 561 | 906 | 10 | 334 | ChainA, Endoglucanase E [Acetivibrio thermocellus],4IM4_B Chain B, Endoglucanase E [Acetivibrio thermocellus],4IM4_C Chain C, Endoglucanase E [Acetivibrio thermocellus],4IM4_D Chain D, Endoglucanase E [Acetivibrio thermocellus],4IM4_E Chain E, Endoglucanase E [Acetivibrio thermocellus],4IM4_F Chain F, Endoglucanase E [Acetivibrio thermocellus] |
| 6MQ4_A | 1.27e-45 | 561 | 907 | 15 | 349 | ChainA, cellulase [Acetivibrio cellulolyticus] |
| 6WQP_A | 1.30e-45 | 550 | 892 | 8 | 348 | GH5-4broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQP_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQV_A GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_C GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_D GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis] |
| 3NDY_A | 5.32e-42 | 554 | 903 | 8 | 344 | Thestructure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_B The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_C The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_D The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDZ_A The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_B The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_C The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_D The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O08342 | 1.25e-46 | 553 | 906 | 35 | 398 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| P23660 | 1.93e-43 | 564 | 875 | 33 | 339 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
| P28621 | 2.41e-43 | 543 | 917 | 26 | 384 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
| P10477 | 2.91e-42 | 561 | 906 | 60 | 384 | Cellulase/esterase CelE OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celE PE=1 SV=2 |
| P16216 | 4.86e-41 | 552 | 892 | 63 | 390 | Endoglucanase 1 OS=Ruminococcus albus OX=1264 GN=Eg I PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
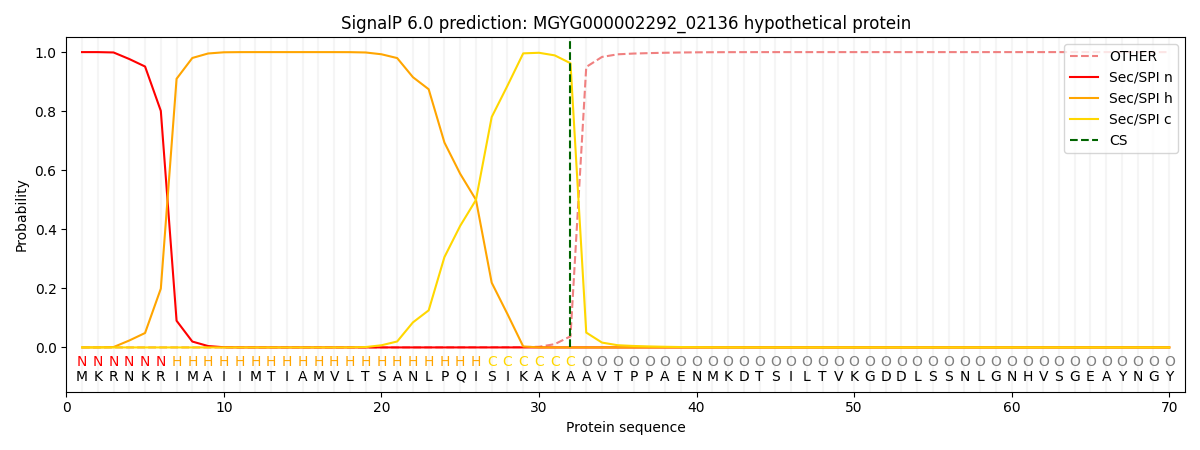
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000222 | 0.999059 | 0.000210 | 0.000186 | 0.000151 | 0.000134 |