You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002295_02325
You are here: Home > Sequence: MGYG000002295_02325
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
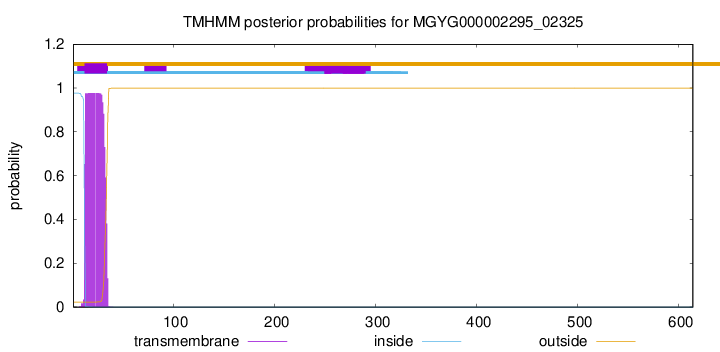
TMHMM annotations
Basic Information help
| Species | Ruminococcus_C callidus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_C; Ruminococcus_C callidus | |||||||||||
| CAZyme ID | MGYG000002295_02325 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 19503; End: 21347 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 134 | 319 | 1.7e-40 | 0.6964980544747081 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.12e-22 | 98 | 374 | 1 | 258 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 4.50e-13 | 94 | 240 | 49 | 191 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| cd14256 | Dockerin_I | 0.005 | 43 | 74 | 18 | 54 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AQG79101.1 | 7.98e-137 | 90 | 610 | 42 | 552 |
| ABG59109.1 | 1.74e-116 | 90 | 609 | 35 | 550 |
| QSI77041.1 | 6.11e-64 | 89 | 330 | 43 | 271 |
| ACR11279.1 | 1.45e-63 | 89 | 342 | 29 | 271 |
| ADX74269.1 | 1.57e-56 | 78 | 609 | 53 | 591 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1CEC_A | 4.79e-23 | 134 | 341 | 35 | 228 | ChainA, ENDOGLUCANASE CELC [Acetivibrio thermocellus] |
| 1CEN_A | 1.18e-22 | 134 | 341 | 35 | 228 | ChainA, CELLULASE CELC [Acetivibrio thermocellus],1CEO_A Chain A, CELLULASE CELC [Acetivibrio thermocellus] |
| 3AMC_A | 6.86e-13 | 136 | 349 | 42 | 236 | Crystalstructures of Thermotoga maritima Cel5A, apo form and dimer/au [Thermotoga maritima MSB8],3AMC_B Crystal structures of Thermotoga maritima Cel5A, apo form and dimer/au [Thermotoga maritima MSB8],3AMD_A Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_B Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_C Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_D Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3MMU_A Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_B Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_C Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_D Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_E Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_F Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_G Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_H Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_A Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_B Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_C Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_D Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima] |
| 3AZR_A | 6.86e-13 | 136 | 349 | 42 | 236 | DiverseSubstrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose [Thermotoga maritima MSB8],3AZR_B Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose [Thermotoga maritima MSB8],3AZS_A Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Mannotriose [Thermotoga maritima MSB8],3AZS_B Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Mannotriose [Thermotoga maritima MSB8],3AZT_A Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose [Thermotoga maritima MSB8],3AZT_B Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose [Thermotoga maritima MSB8],3AZT_C Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose [Thermotoga maritima MSB8],3AZT_D Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose [Thermotoga maritima MSB8] |
| 3AMG_A | 3.97e-12 | 136 | 349 | 42 | 236 | Crystalstructures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form [Thermotoga maritima MSB8],3AMG_B Crystal structures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form [Thermotoga maritima MSB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23340 | 1.07e-22 | 134 | 341 | 35 | 228 | Endoglucanase C307 OS=Clostridium sp. (strain F1) OX=1508 GN=celC307 PE=1 SV=1 |
| A3DJ77 | 1.07e-22 | 134 | 341 | 35 | 228 | Endoglucanase C OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celC PE=3 SV=1 |
| P0C2S3 | 2.62e-22 | 134 | 341 | 35 | 228 | Endoglucanase C OS=Acetivibrio thermocellus OX=1515 GN=celC PE=1 SV=1 |
| P14250 | 2.35e-15 | 98 | 374 | 302 | 561 | Endoglucanase 3 OS=Fibrobacter succinogenes (strain ATCC 19169 / S85) OX=59374 GN=cel-3 PE=1 SV=2 |
| P16169 | 4.18e-14 | 134 | 360 | 37 | 258 | Cellodextrinase A OS=Ruminococcus flavefaciens OX=1265 GN=celA PE=3 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
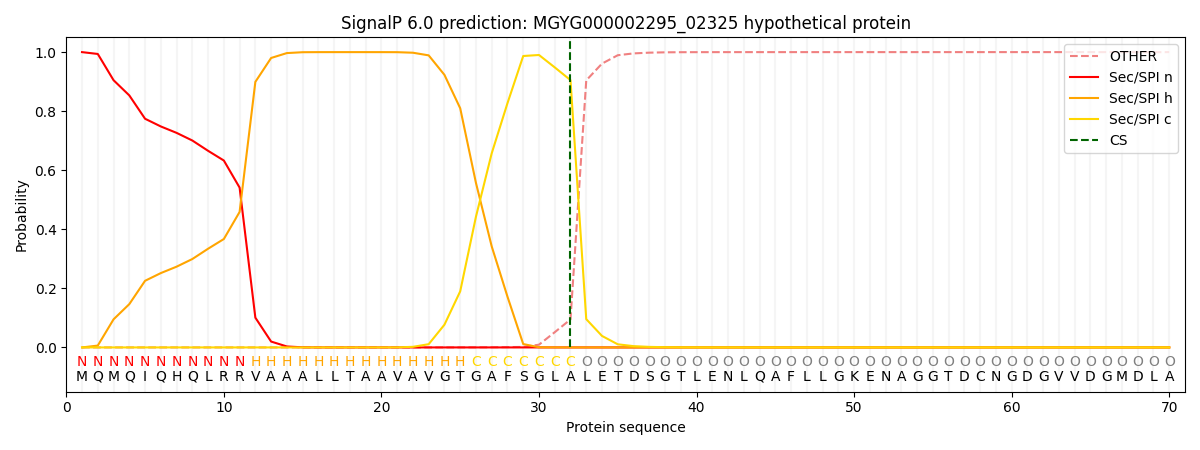
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000694 | 0.998504 | 0.000214 | 0.000197 | 0.000182 | 0.000168 |