You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002300_00013
You are here: Home > Sequence: MGYG000002300_00013
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides cutis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides cutis | |||||||||||
| CAZyme ID | MGYG000002300_00013 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16013; End: 17758 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 332 | 572 | 9.4e-32 | 0.6765676567656765 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 9.17e-15 | 322 | 571 | 44 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.96e-08 | 339 | 573 | 126 | 339 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIU93949.1 | 1.47e-128 | 1 | 577 | 1 | 727 |
| QRQ48288.1 | 6.33e-127 | 1 | 575 | 3 | 716 |
| QDM11106.1 | 1.23e-126 | 1 | 577 | 1 | 727 |
| QUR44907.1 | 7.04e-126 | 1 | 577 | 1 | 727 |
| QGT73069.1 | 1.40e-125 | 1 | 577 | 1 | 727 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6LPS_A | 1.13e-09 | 326 | 565 | 113 | 323 | ChainA, Beta-xylanase [Halalkalibacterium halodurans] |
| 2UWF_A | 3.64e-09 | 326 | 565 | 112 | 322 | ChainA, ALKALINE ACTIVE ENDOXYLANASE [Halalkalibacterium halodurans] |
| 7CPL_A | 3.71e-09 | 326 | 565 | 114 | 324 | XylanaseR from Bacillus sp. TAR-1 [Bacillus sp. TAR1] |
| 7CPK_A | 3.71e-09 | 326 | 565 | 114 | 324 | XylanaseR from Bacillus sp. TAR-1 [Bacillus sp. TAR1] |
| 2F8Q_A | 1.40e-06 | 340 | 565 | 125 | 325 | Analkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27],2F8Q_B An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P07528 | 2.35e-08 | 326 | 565 | 158 | 368 | Endo-1,4-beta-xylanase A OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=xynA PE=1 SV=1 |
| Q9HEZ1 | 2.92e-06 | 318 | 575 | 178 | 406 | Endo-1,4-beta-xylanase A OS=Phanerodontia chrysosporium OX=2822231 GN=xynA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
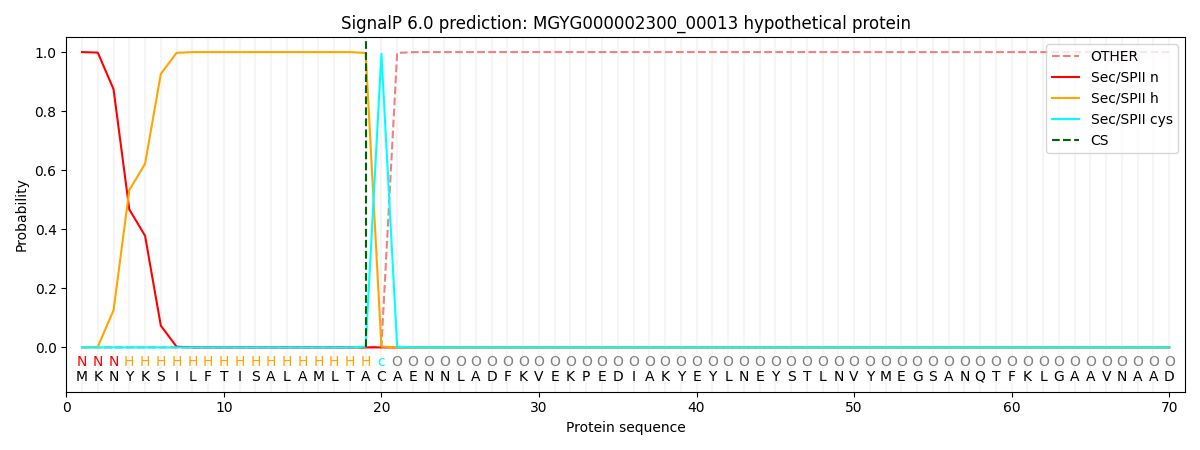
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000038 | 0.000000 | 0.000000 | 0.000000 |
