You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002300_00187
You are here: Home > Sequence: MGYG000002300_00187
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides cutis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides cutis | |||||||||||
| CAZyme ID | MGYG000002300_00187 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 235317; End: 236489 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 252 | 384 | 5.1e-18 | 0.967741935483871 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16389 | DUF4998 | 1.85e-48 | 20 | 209 | 1 | 195 | Domain of unknown function. This family around 200 residues locates in the N-terminal of some uncharacterized proteins in various Bacteroides and Parabacteroides species. The function of this family remains unknown. |
| pfam00754 | F5_F8_type_C | 1.78e-16 | 251 | 383 | 2 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 7.15e-06 | 251 | 376 | 14 | 135 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMW86484.1 | 9.54e-148 | 1 | 386 | 1 | 388 |
| BCA48665.1 | 5.47e-147 | 1 | 386 | 1 | 388 |
| QUT41988.1 | 1.33e-142 | 15 | 386 | 4 | 377 |
| QQT78590.1 | 9.68e-133 | 1 | 386 | 1 | 388 |
| QRP59525.1 | 9.68e-133 | 1 | 386 | 1 | 388 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2KD7_A | 3.65e-22 | 244 | 383 | 5 | 140 | ChainA, Putative chitobiase [Bacteroides thetaiotaomicron VPI-5482] |
| 3F2Z_A | 3.65e-22 | 242 | 383 | 3 | 143 | Crystalstructure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B [Bacteroides fragilis NCTC 9343] |
| 3GGL_A | 3.26e-21 | 244 | 383 | 15 | 150 | X-RayStructure of the C-terminal domain (277-440) of Putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR324A. [Bacteroides thetaiotaomicron],6OE2_A X-Ray Structure of the C-terminal domain (277-440) of Putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR324A. Re-refinement of 3GGL with correct metal Mn replacing Zn. New metal confirmed with PIXE analysis of original sample. [Bacteroides thetaiotaomicron] |
| 2BZD_A | 1.07e-06 | 265 | 383 | 477 | 595 | Galactoserecognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_B Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_C Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens] |
| 2BER_A | 1.07e-06 | 265 | 383 | 477 | 595 | Y370GActive Site Mutant of the Sialidase from Micromonospora viridifaciens in complex with beta-Neu5Ac (sialic acid). [Micromonospora viridifaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| I1S2N3 | 4.64e-06 | 244 | 388 | 50 | 195 | Galactose oxidase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=GAOA PE=3 SV=1 |
| Q02834 | 6.02e-06 | 265 | 383 | 523 | 641 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| P0CS93 | 6.14e-06 | 244 | 388 | 50 | 195 | Galactose oxidase OS=Gibberella zeae OX=5518 GN=GAOA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
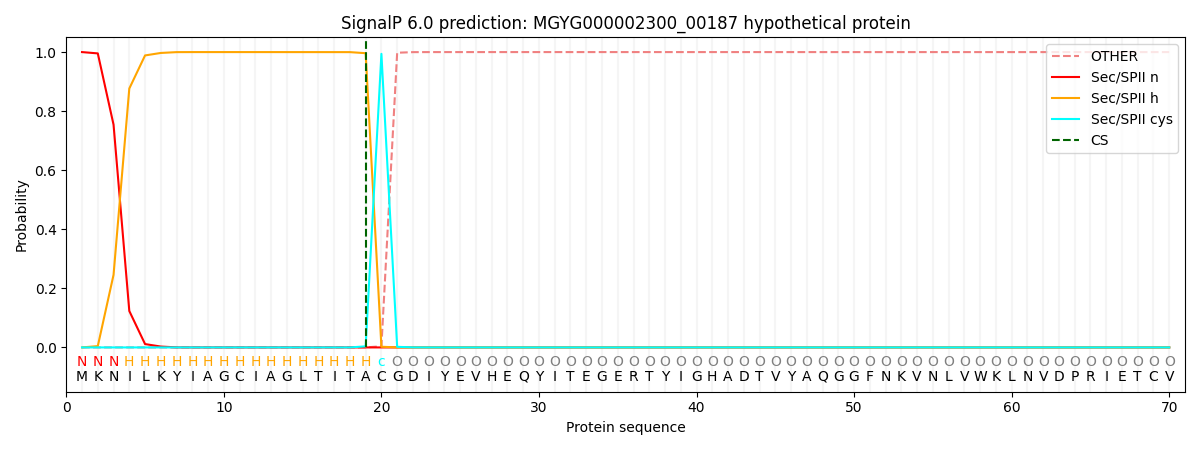
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000049 | 0.000000 | 0.000000 | 0.000000 |
