You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002300_00964
You are here: Home > Sequence: MGYG000002300_00964
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
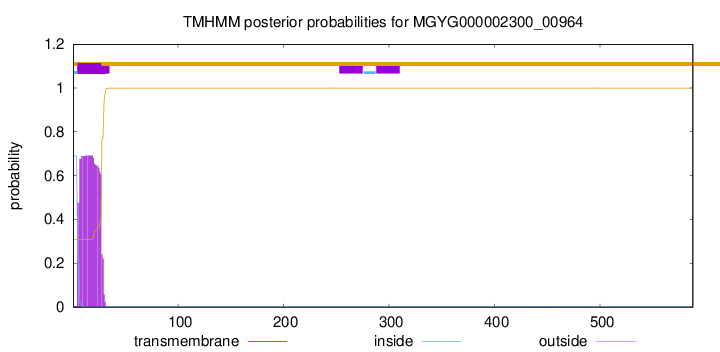
TMHMM annotations
Basic Information help
| Species | Bacteroides cutis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides cutis | |||||||||||
| CAZyme ID | MGYG000002300_00964 | |||||||||||
| CAZy Family | CE12 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 233462; End: 235228 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 286 | 558 | 2.3e-100 | 0.9444444444444444 |
| CE12 | 26 | 270 | 7.4e-79 | 0.9952380952380953 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01821 | Rhamnogalacturan_acetylesterase_like | 2.83e-88 | 26 | 270 | 2 | 198 | Rhamnogalacturan_acetylesterase_like subgroup of SGNH-hydrolases. Rhamnogalacturan acetylesterase removes acetyl esters from rhamnogalacturonan substrates, and renders them susceptible to degradation by rhamnogalacturonases. Rhamnogalacturonans are highly branched regions in pectic polysaccharides, consisting of repeating -(1,2)-L-Rha-(1,4)-D-GalUA disaccharide units, with many rhamnose residues substituted by neutral oligosaccharides such as arabinans, galactans and arabinogalactans. Extracellular enzymes participating in the degradation of plant cell wall polymers, such as Rhamnogalacturonan acetylesterase, would typically be found in saprophytic and plant pathogenic fungi and bacteria. |
| PLN02773 | PLN02773 | 5.25e-83 | 288 | 569 | 9 | 292 | pectinesterase |
| pfam01095 | Pectinesterase | 7.76e-79 | 285 | 549 | 1 | 271 | Pectinesterase. |
| PLN02682 | PLN02682 | 3.95e-69 | 284 | 556 | 69 | 347 | pectinesterase family protein |
| COG4677 | PemB | 7.03e-69 | 285 | 543 | 81 | 378 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT45771.1 | 0.0 | 1 | 588 | 1 | 588 |
| QRQ48636.1 | 0.0 | 1 | 588 | 1 | 588 |
| ALJ61265.1 | 0.0 | 1 | 588 | 1 | 588 |
| QUT93206.1 | 0.0 | 1 | 588 | 1 | 588 |
| QDO69180.1 | 0.0 | 1 | 588 | 1 | 588 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1XG2_A | 3.58e-45 | 287 | 556 | 6 | 283 | ChainA, Pectinesterase 1 [Solanum lycopersicum] |
| 1GQ8_A | 1.00e-44 | 287 | 576 | 10 | 298 | Pectinmethylesterase from Carrot [Daucus carota] |
| 2NSP_A | 3.47e-34 | 284 | 577 | 4 | 340 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 3UW0_A | 5.38e-34 | 284 | 558 | 31 | 351 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
| 5C1E_A | 1.40e-31 | 294 | 549 | 17 | 271 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9LVQ0 | 1.98e-61 | 288 | 556 | 9 | 282 | Pectinesterase 31 OS=Arabidopsis thaliana OX=3702 GN=PME31 PE=1 SV=1 |
| Q8VYZ3 | 2.08e-50 | 290 | 556 | 90 | 361 | Probable pectinesterase 53 OS=Arabidopsis thaliana OX=3702 GN=PME53 PE=2 SV=1 |
| P41510 | 1.98e-47 | 287 | 567 | 274 | 563 | Probable pectinesterase/pectinesterase inhibitor OS=Brassica napus OX=3708 GN=BP19 PE=2 SV=1 |
| Q43143 | 4.68e-46 | 285 | 566 | 272 | 552 | Pectinesterase/pectinesterase inhibitor U1 OS=Solanum lycopersicum OX=4081 GN=PMEU1 PE=2 SV=1 |
| Q1JPL7 | 5.98e-46 | 287 | 549 | 248 | 516 | Pectinesterase/pectinesterase inhibitor 18 OS=Arabidopsis thaliana OX=3702 GN=PME18 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
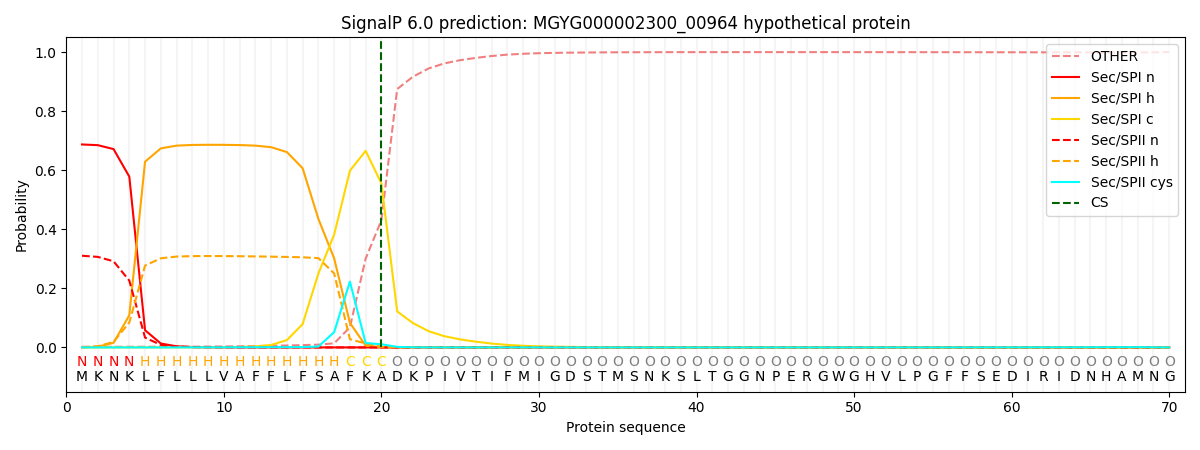
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003824 | 0.678829 | 0.316519 | 0.000279 | 0.000269 | 0.000261 |