You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002306_01884
You are here: Home > Sequence: MGYG000002306_01884
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
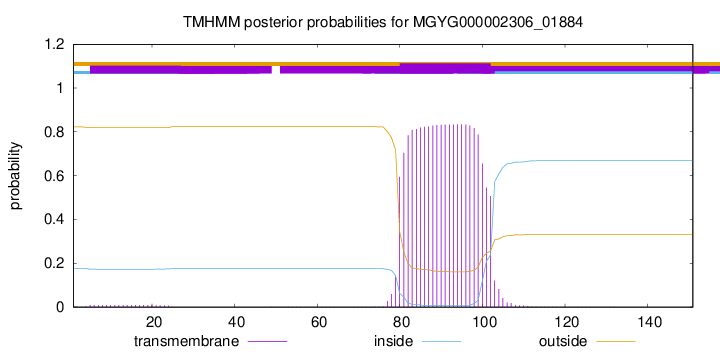
TMHMM annotations
Basic Information help
| Species | Mycobacterium avium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Mycobacterium; Mycobacterium avium | |||||||||||
| CAZyme ID | MGYG000002306_01884 | |||||||||||
| CAZy Family | GT1 | |||||||||||
| CAZyme Description | UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3905; End: 4360 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08660 | Alg14 | 1.03e-22 | 4 | 150 | 1 | 171 | Oligosaccharide biosynthesis protein Alg14 like. Alg14 is involved dolichol-linked oligosaccharide biosynthesis and anchors the catalytic subunit Alg13 to the ER membrane. |
| COG0707 | MurG | 1.56e-12 | 2 | 144 | 1 | 154 | UDP-N-acetylglucosamine:LPS N-acetylglucosamine transferase [Cell wall/membrane/envelope biogenesis]. |
| cd03785 | GT28_MurG | 7.37e-07 | 63 | 144 | 74 | 152 | undecaprenyldiphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase. MurG (EC 2.4.1.227) is an N-acetylglucosaminyltransferase, the last enzyme involved in the intracellular phase of peptidoglycan biosynthesis. It transfers N-acetyl-D-glucosamine (GlcNAc) from UDP-GlcNAc to the C4 hydroxyl of a lipid-linked N-acetylmuramoyl pentapeptide (NAM). The resulting disaccharide is then transported across the cell membrane, where it is polymerized into NAG-NAM cell-wall repeat structure. MurG belongs to the GT-B structural superfamily of glycoslytransferases, which have characteristic N- and C-terminal domains, each containing a typical Rossmann fold. The two domains have high structural homology despite minimal sequence homology. The large cleft that separates the two domains includes the catalytic center and permits a high degree of flexibility. |
| PRK00726 | murG | 1.32e-04 | 57 | 144 | 70 | 154 | undecaprenyldiphospho-muramoylpentapeptide beta-N- acetylglucosaminyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AJK80256.1 | 3.89e-106 | 1 | 151 | 1 | 151 |
| ATO70861.1 | 3.89e-106 | 1 | 151 | 1 | 151 |
| AYQ69611.1 | 3.89e-106 | 1 | 151 | 1 | 151 |
| AJK76063.1 | 3.89e-106 | 1 | 151 | 1 | 151 |
| QPM72238.1 | 3.89e-106 | 1 | 151 | 1 | 151 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q6CF02 | 8.46e-14 | 66 | 151 | 125 | 216 | UDP-N-acetylglucosamine transferase subunit ALG14 OS=Yarrowia lipolytica (strain CLIB 122 / E 150) OX=284591 GN=ALG14 PE=3 SV=1 |
| Q5A5N6 | 8.84e-10 | 3 | 151 | 49 | 219 | UDP-N-acetylglucosamine transferase subunit ALG14 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=ALG14 PE=3 SV=1 |
| Q9D081 | 1.66e-09 | 3 | 151 | 38 | 217 | UDP-N-acetylglucosamine transferase subunit ALG14 homolog OS=Mus musculus OX=10090 GN=Alg14 PE=1 SV=1 |
| Q6AY85 | 1.15e-08 | 36 | 151 | 95 | 216 | UDP-N-acetylglucosamine transferase subunit ALG14 homolog OS=Rattus norvegicus OX=10116 GN=Alg14 PE=2 SV=1 |
| Q96F25 | 2.19e-08 | 59 | 151 | 118 | 216 | UDP-N-acetylglucosamine transferase subunit ALG14 homolog OS=Homo sapiens OX=9606 GN=ALG14 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000053 | 0.000003 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |