You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002306_02021
You are here: Home > Sequence: MGYG000002306_02021
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Mycobacterium avium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Mycobacterium; Mycobacterium avium | |||||||||||
| CAZyme ID | MGYG000002306_02021 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 359; End: 1021 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd13402 | LT_TF-like | 1.68e-11 | 116 | 216 | 1 | 92 | lytic transglycosylase-like domain of tail fiber-like proteins and similar domains. These tail fiber-like proteins are multi-domain proteins that include a lytic transglycosylase (LT) domain. Members of the LT family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, and the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| COG3953 | SLT | 1.34e-08 | 115 | 197 | 14 | 89 | SLT domain protein [Mobilome: prophages, transposons]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBN49321.1 | 5.94e-162 | 1 | 220 | 1 | 220 |
| QKU44270.1 | 5.94e-162 | 1 | 220 | 1 | 220 |
| BAN32317.1 | 5.94e-162 | 1 | 220 | 1 | 220 |
| QGW31131.1 | 5.94e-162 | 1 | 220 | 1 | 220 |
| AGL35741.1 | 2.19e-161 | 1 | 220 | 37 | 256 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P45931 | 8.84e-09 | 115 | 216 | 1363 | 1461 | Uncharacterized protein YqbO OS=Bacillus subtilis (strain 168) OX=224308 GN=yqbO PE=1 SV=2 |
| P54334 | 4.20e-07 | 115 | 216 | 1111 | 1209 | Phage-like element PBSX protein XkdO OS=Bacillus subtilis (strain 168) OX=224308 GN=xkdO PE=4 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
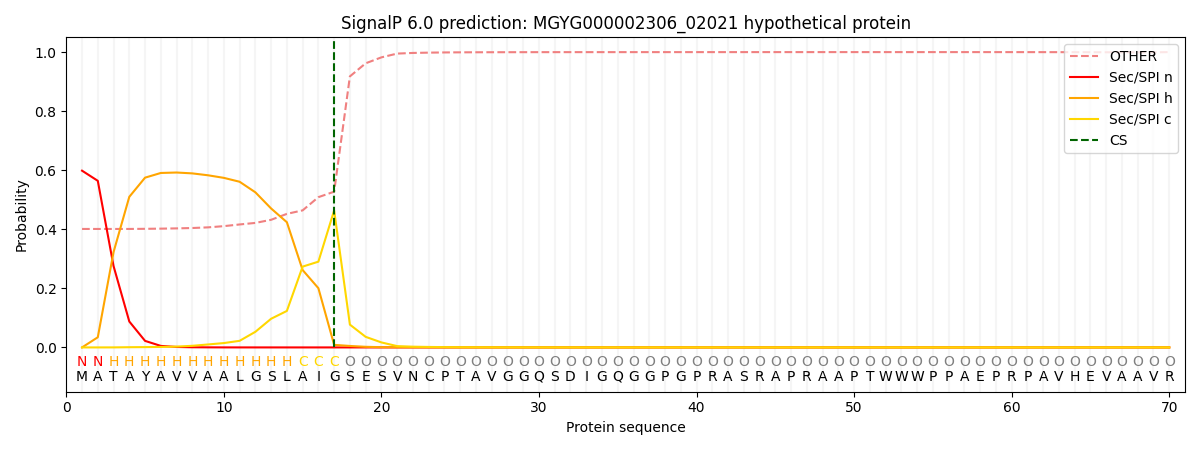
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.413117 | 0.585120 | 0.000975 | 0.000288 | 0.000231 | 0.000268 |
