You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002306_03165
You are here: Home > Sequence: MGYG000002306_03165
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
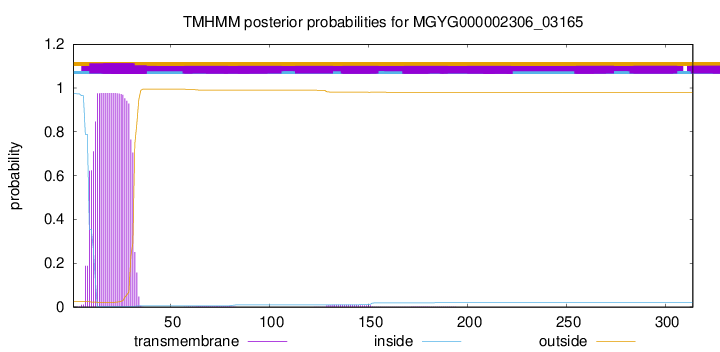
TMHMM annotations
Basic Information help
| Species | Mycobacterium avium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Mycobacterium; Mycobacterium avium | |||||||||||
| CAZyme ID | MGYG000002306_03165 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3716; End: 4660 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 52 | 203 | 4.7e-24 | 0.6079295154185022 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3509 | LpqC | 8.44e-66 | 44 | 314 | 36 | 309 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| pfam10503 | Esterase_phd | 4.51e-14 | 56 | 256 | 2 | 216 | Esterase PHB depolymerase. This family of proteins include acetyl xylan esterases (AXE), feruloyl esterases (FAE), and poly(3-hydroxybutyrate) (PHB) depolymerases. |
| COG0412 | DLH | 3.41e-12 | 59 | 206 | 18 | 173 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
| COG1506 | DAP2 | 1.71e-11 | 56 | 206 | 379 | 566 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| TIGR01840 | esterase_phb | 4.82e-09 | 57 | 180 | 1 | 128 | esterase, PHB depolymerase family. This model describes a subfamily among lipases of the ab-hydrolase family. This subfamily includes bacterial depolymerases for poly(3-hydroxybutyrate) (PHB) and related polyhydroxyalkanoates (PHA), as well as acetyl xylan esterases, feruloyl esterases, and others from fungi. [Fatty acid and phospholipid metabolism, Degradation] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AIE87169.1 | 4.82e-28 | 54 | 309 | 10 | 257 |
| ALO15718.1 | 6.43e-24 | 52 | 311 | 32 | 288 |
| ACL94355.2 | 2.09e-22 | 6 | 314 | 12 | 303 |
| ACX75364.1 | 4.03e-20 | 48 | 277 | 30 | 241 |
| ADL27269.1 | 4.03e-20 | 48 | 277 | 30 | 241 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8YG19 | 5.73e-14 | 52 | 207 | 55 | 205 | Bifunctional acetylxylan esterase/xylanase XynS20E OS=Neocallimastix patriciarum OX=4758 GN=xynS20E PE=1 SV=1 |
| Q5B2G3 | 3.93e-11 | 13 | 309 | 6 | 267 | Feruloyl esterase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=faeC PE=1 SV=1 |
| Q0CDX2 | 6.01e-10 | 55 | 309 | 43 | 267 | Probable feruloyl esterase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=faeC PE=3 SV=1 |
| A1C9D4 | 5.00e-09 | 66 | 309 | 58 | 269 | Probable feruloyl esterase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=faeC-1 PE=3 SV=1 |
| A2QYU7 | 3.98e-08 | 55 | 309 | 43 | 267 | Probable feruloyl esterase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=faeC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
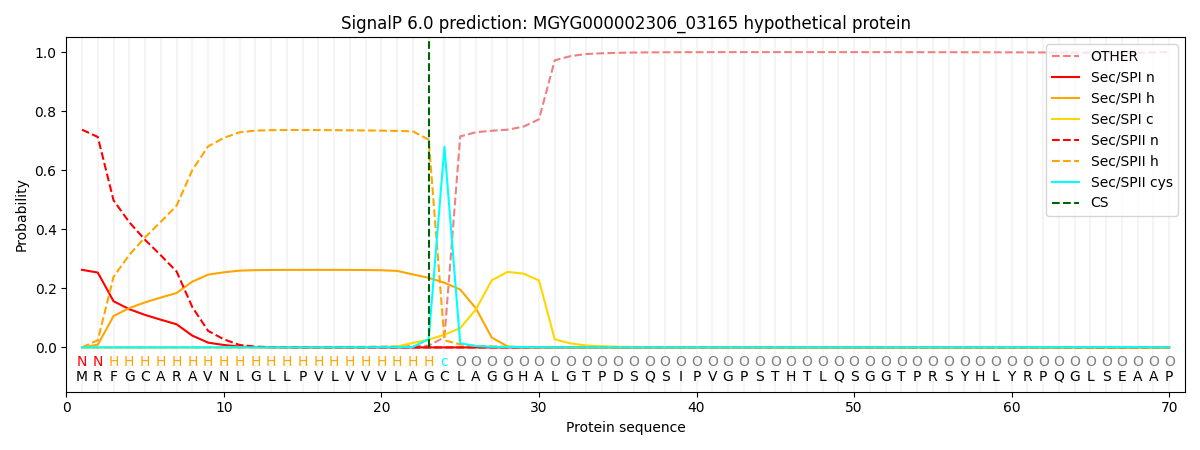
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000443 | 0.256250 | 0.742623 | 0.000305 | 0.000235 | 0.000147 |