You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002306_03770
Basic Information
help
| Species |
Mycobacterium avium
|
| Lineage |
Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Mycobacterium; Mycobacterium avium
|
| CAZyme ID |
MGYG000002306_03770
|
| CAZy Family |
GT83 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000002306 |
4669710 |
Isolate |
Australia |
Oceania |
|
| Gene Location |
Start: 7261;
End: 8838
Strand: -
|
No EC number prediction in MGYG000002306_03770.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
84 |
387 |
2.3e-35 |
0.5518518518518518 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG5305
|
COG5305 |
2.44e-101 |
25 |
524 |
43 |
551 |
Uncharacterized membrane protein [Function unknown]. |
This protein is predicted as OTHER
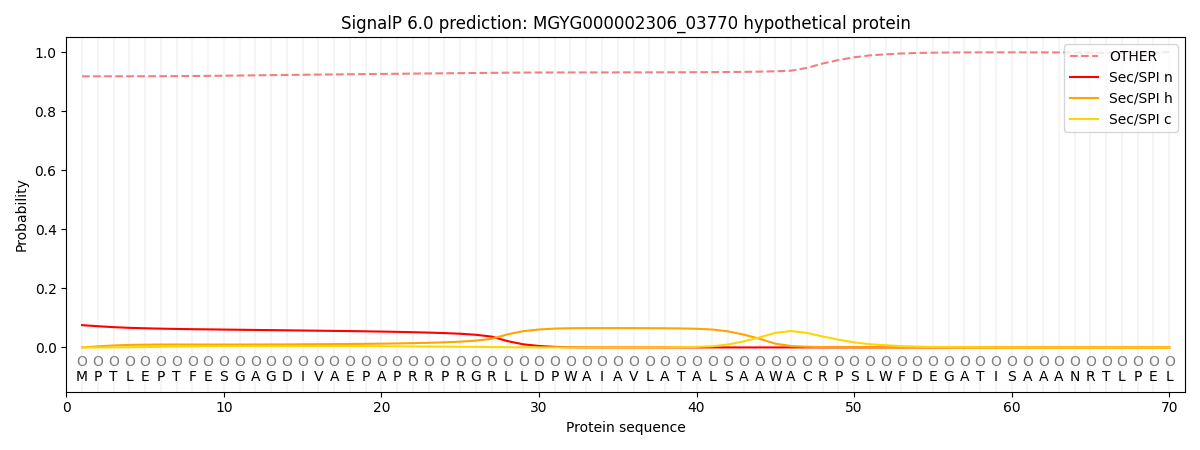
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.921940
|
0.069124
|
0.006116
|
0.000415
|
0.000273
|
0.002135
|

| start |
end |
| 153 |
175 |
| 182 |
204 |
| 219 |
241 |
| 271 |
293 |
| 303 |
325 |
| 332 |
354 |
| 358 |
377 |


