You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002306_04057
You are here: Home > Sequence: MGYG000002306_04057
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Mycobacterium avium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Mycobacterium; Mycobacterium avium | |||||||||||
| CAZyme ID | MGYG000002306_04057 | |||||||||||
| CAZy Family | CE5 | |||||||||||
| CAZyme Description | putative cutinase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 482; End: 1180 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 22 | 213 | 8.5e-62 | 0.9947089947089947 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01083 | Cutinase | 6.10e-44 | 22 | 213 | 1 | 173 | Cutinase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBN50642.1 | 8.06e-166 | 1 | 232 | 1 | 232 |
| BAN33618.1 | 8.06e-166 | 1 | 232 | 1 | 232 |
| QGW34718.1 | 8.06e-166 | 1 | 232 | 1 | 232 |
| QQK52207.1 | 1.39e-165 | 1 | 232 | 16 | 247 |
| AYQ67943.1 | 1.39e-165 | 1 | 232 | 16 | 247 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4PSC_A | 5.38e-18 | 23 | 198 | 79 | 235 | Structureof cutinase from Trichoderma reesei in its native form. [Trichoderma reesei QM6a],4PSD_A Structure of Trichoderma reesei cutinase native form. [Trichoderma reesei QM6a],4PSE_A Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a],4PSE_B Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a] |
| 7CY3_A | 6.23e-11 | 20 | 198 | 14 | 180 | Crystalstructure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9. [Paraphoma sp. B47-9],7CY3_B Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9. [Paraphoma sp. B47-9],7CY9_A Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9 solved by getting the phase from anomalous scattering of uncovalently coordinated arsenic (cacodylate). [Paraphoma sp. B47-9],7CY9_B Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9 solved by getting the phase from anomalous scattering of uncovalently coordinated arsenic (cacodylate). [Paraphoma sp. B47-9] |
| 2AXE_A | 4.49e-09 | 23 | 214 | 2 | 207 | IodinatedComplex Of Acetyl Xylan Esterase At 1.80 Angstroms [Talaromyces purpureogenus] |
| 1BS9_A | 6.14e-09 | 23 | 214 | 2 | 207 | AcetylxylanEsterase From P. Purpurogenum Refined At 1.10 Angstroms [Talaromyces purpureogenus] |
| 1G66_A | 6.14e-09 | 23 | 214 | 2 | 207 | ACETYLXYLANESTERASE AT 0.90 ANGSTROM RESOLUTION [Talaromyces purpureogenus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O06793 | 3.60e-61 | 47 | 214 | 9 | 174 | Probable carboxylesterase Culp5 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=cut1 PE=3 SV=1 |
| Q79FA4 | 6.46e-52 | 54 | 213 | 23 | 187 | Probable carboxylesterase Culp7 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=cut5b PE=2 SV=1 |
| P9WP43 | 2.38e-45 | 23 | 213 | 35 | 213 | Carboxylesterase Culp1 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=cut7 PE=1 SV=1 |
| P63880 | 2.38e-45 | 23 | 213 | 35 | 213 | Carboxylesterase Culp1 OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=BQ2027_MB2006C PE=3 SV=1 |
| P9WP42 | 2.38e-45 | 23 | 213 | 35 | 213 | Carboxylesterase Culp1 OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=MT2037 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
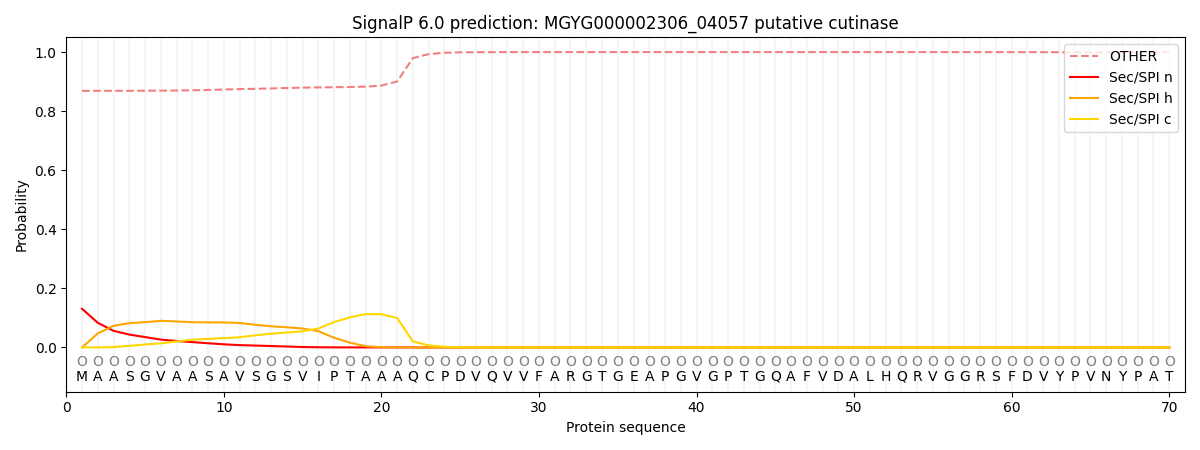
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.874938 | 0.123953 | 0.000510 | 0.000307 | 0.000130 | 0.000170 |
