You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002310_00567
You are here: Home > Sequence: MGYG000002310_00567
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Microvirga massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Beijerinckiaceae; Microvirga; Microvirga massiliensis | |||||||||||
| CAZyme ID | MGYG000002310_00567 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 574596; End: 575615 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 152 | 285 | 1.2e-26 | 0.40924092409240925 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4124 | ManB2 | 8.92e-91 | 73 | 335 | 80 | 348 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| pfam02156 | Glyco_hydro_26 | 5.24e-08 | 156 | 283 | 136 | 273 | Glycosyl hydrolase family 26. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIG52731.1 | 1.37e-124 | 55 | 333 | 24 | 304 |
| AYG61987.1 | 8.40e-123 | 64 | 337 | 44 | 317 |
| ATU94264.1 | 3.40e-122 | 36 | 336 | 21 | 316 |
| QYA16675.1 | 7.42e-118 | 32 | 334 | 16 | 309 |
| QUE83811.1 | 1.49e-117 | 66 | 333 | 41 | 308 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2DDX_A | 4.94e-10 | 145 | 260 | 84 | 201 | Crystalstructure of beta-1,3-xylanase from Vibrio sp. AX-4 [Vibrio sp. AX-4],3VPL_A Crystal structure of a 2-fluoroxylotriosyl complex of the Vibrio sp. AX-4 Beta-1,3-xylanase [Vibrio sp.] |
| 1J9Y_A | 1.08e-06 | 64 | 339 | 44 | 348 | Crystalstructure of mannanase 26A from Pseudomonas cellulosa [Cellvibrio japonicus] |
| 1R7O_A | 1.10e-06 | 64 | 339 | 54 | 358 | CrystalStructure of apo-mannanase 26A from Psudomonas cellulosa [Cellvibrio japonicus] |
| 1GVY_A | 4.48e-06 | 64 | 339 | 44 | 348 | Substratedistorsion by beta-mannanase from Pseudomonas cellulosa [Cellvibrio japonicus] |
| 1ODZ_A | 7.98e-06 | 64 | 339 | 45 | 349 | Expansionof the glycosynthase repertoire to produce defined manno-oligosaccharides [Cellvibrio japonicus],1ODZ_B Expansion of the glycosynthase repertoire to produce defined manno-oligosaccharides [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5MP61 | 4.48e-09 | 145 | 260 | 106 | 223 | Beta-1,3-xylanase XYL4 OS=Vibrio sp. OX=678 GN=xyl4 PE=1 SV=1 |
| Q8RS40 | 3.82e-06 | 147 | 260 | 108 | 223 | Beta-1,3-xylanase OS=Alcaligenes sp. OX=512 GN=txyA PE=1 SV=1 |
| P49424 | 6.33e-06 | 64 | 339 | 82 | 386 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=manA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
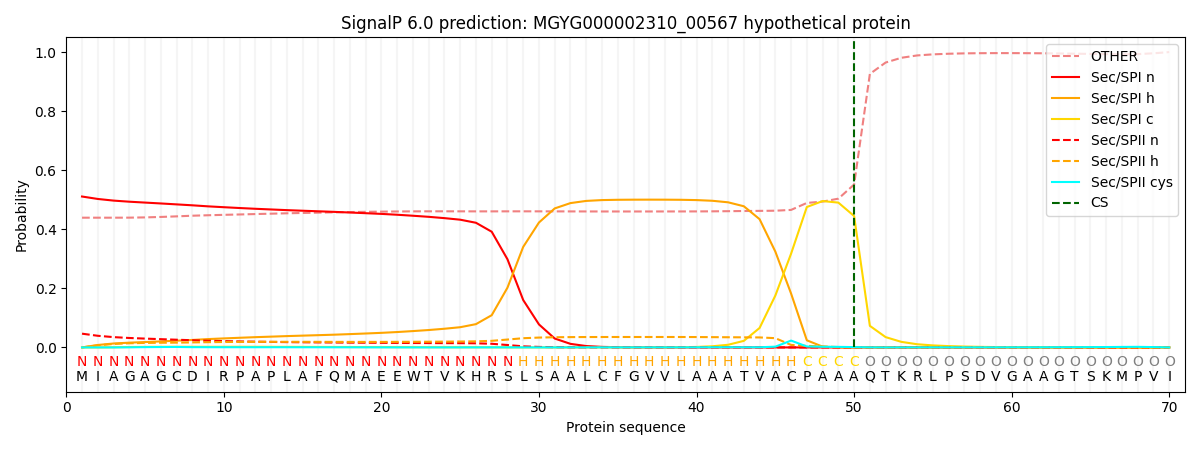
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.449927 | 0.487794 | 0.056026 | 0.003691 | 0.001674 | 0.000885 |
