You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002311_03433
You are here: Home > Sequence: MGYG000002311_03433
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Vibrio cholerae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Vibrionaceae; Vibrio; Vibrio cholerae | |||||||||||
| CAZyme ID | MGYG000002311_03433 | |||||||||||
| CAZy Family | AA10 | |||||||||||
| CAZyme Description | GlcNAc-binding protein A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 786538; End: 787707 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA10 | 26 | 214 | 9.1e-50 | 0.9887640449438202 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3397 | COG3397 | 4.83e-110 | 4 | 297 | 10 | 282 | Predicted carbohydrate-binding protein, contains CBM5 and CBM33 domains [General function prediction only]. |
| pfam03067 | LPMO_10 | 5.52e-60 | 26 | 214 | 1 | 186 | Lytic polysaccharide mono-oxygenase, cellulose-degrading. This domain is found associated with a wide variety of cellulose binding domains. This is a family of two very closely related proteins that together act as both a C1- and a C4-oxidising lytic polysaccharide mono-oxygenase, degrading cellulose. This domain is also found in baculoviral spheroidins and spindolins, protein of unknown function. |
| cd21177 | LPMO_AA10 | 4.23e-57 | 26 | 215 | 1 | 180 | lytic polysaccharide monooxygenase (LPMO) auxiliary activity family 10 (AA10). AA10 proteins are copper-dependent lytic polysaccharide monooxygenases (LPMOs), which may act on chitin or cellulose. The family used to be called CBM33. Activities in this family include lytic cellulose monooxygenase (C1-hydroxylating) (EC 1.14.99.54), lytic cellulose monooxygenase (C4-dehydrogenating) (EC 1.14.99.56), lytic chitin monooxygenase (EC 1.14.99.53), and lytic xylan monooxygenase/xylan oxidase (glycosidic bond-cleaving) (EC 1.14.99.-). Also included are viral chitin-binding glycoproteins such as fusolin and spheroidin-like proteins. |
| PHA03387 | gp37 | 6.60e-49 | 13 | 216 | 7 | 250 | spherodin-like protein; Provisional |
| cd21178 | Fusolin-like | 7.99e-48 | 26 | 216 | 1 | 227 | fusolin and similar proteins. Fusolin is a protein found in spindles of insect poxviruses that resembles the lytic polysaccharide monooxygenases of chitinovorous bacteria and may function to disrupt the chitin-rich peritrophic matrix that protects insects against oral infections. Thus, it is a component of the virus occlusion bodies (which are large proteinaceous polyhedra) that protect the virus from the outside environment for extended periods until they are ingested by insect larvae. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUY00890.1 | 1.13e-294 | 1 | 389 | 1 | 389 |
| APF54288.1 | 1.13e-294 | 1 | 389 | 1 | 389 |
| APF77551.1 | 1.13e-294 | 1 | 389 | 1 | 389 |
| APF61823.1 | 1.13e-294 | 1 | 389 | 1 | 389 |
| ACQ62076.1 | 1.13e-294 | 1 | 389 | 1 | 389 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4YN1_A | 6.49e-25 | 26 | 225 | 1 | 241 | THEATOMIC STRUCTURE OF ANOMALA CUPREA ENTOMOPOXVIRUS (ACEPV) FUSOLIN SPINDLES [Anomala cuprea entomopoxvirus] |
| 4YN2_A | 1.09e-23 | 26 | 224 | 1 | 238 | THEATOMIC STRUCTURE OF WISEANA SPP ENTOMOPOXVIRUS (WSEPV) FUSOLIN SPINDLES [unidentified entomopoxvirus] |
| 4OY7_A | 2.46e-22 | 26 | 218 | 1 | 195 | Structureof cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. [Streptomyces coelicolor A3(2)],4OY7_B Structure of cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. [Streptomyces coelicolor A3(2)],4OY7_C Structure of cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. [Streptomyces coelicolor A3(2)],4OY7_D Structure of cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. [Streptomyces coelicolor A3(2)],4OY7_E Structure of cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. [Streptomyces coelicolor A3(2)],4OY7_F Structure of cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. [Streptomyces coelicolor A3(2)],4OY7_G Structure of cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. [Streptomyces coelicolor A3(2)],4OY7_H Structure of cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. [Streptomyces coelicolor A3(2)] |
| 4X27_A | 8.69e-21 | 26 | 216 | 1 | 233 | Structuralbasis for the enhancement of virulence by entomopoxvirus fusolin and its in vivo crystallization into viral spindles (complex with Copper) [Entomopoxvirinae],4X29_A Structural basis for the enhancement of virulence by entomopoxvirus fusolin and its in vivo crystallization into viral spindles (complex with Zinc) [Entomopoxvirinae] |
| 4OW5_A | 8.78e-21 | 26 | 216 | 1 | 233 | Structuralbasis for the enhancement of virulence by entomopoxvirus fusolin and its in vivo crystallization into viral spindles [unidentified entomopoxvirus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9I589 | 2.74e-34 | 1 | 291 | 1 | 274 | Chitin-binding protein CbpD OS=Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) OX=208964 GN=cbpD PE=1 SV=1 |
| Q02I11 | 1.40e-33 | 1 | 291 | 1 | 274 | Chitin-binding protein CbpD OS=Pseudomonas aeruginosa (strain UCBPP-PA14) OX=208963 GN=cpbD PE=1 SV=1 |
| Q65328 | 3.67e-30 | 14 | 217 | 7 | 249 | Spheroidin-like protein OS=Orgyia pseudotsugata multicapsid polyhedrosis virus OX=262177 GN=SLP PE=2 SV=1 |
| P23058 | 2.47e-29 | 9 | 216 | 3 | 249 | Spheroidin-like protein OS=Autographa californica nuclear polyhedrosis virus OX=46015 GN=SLP PE=2 SV=1 |
| Q05894 | 3.81e-26 | 23 | 224 | 18 | 261 | Spindolin OS=Heliothis armigera entomopoxvirus OX=10290 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
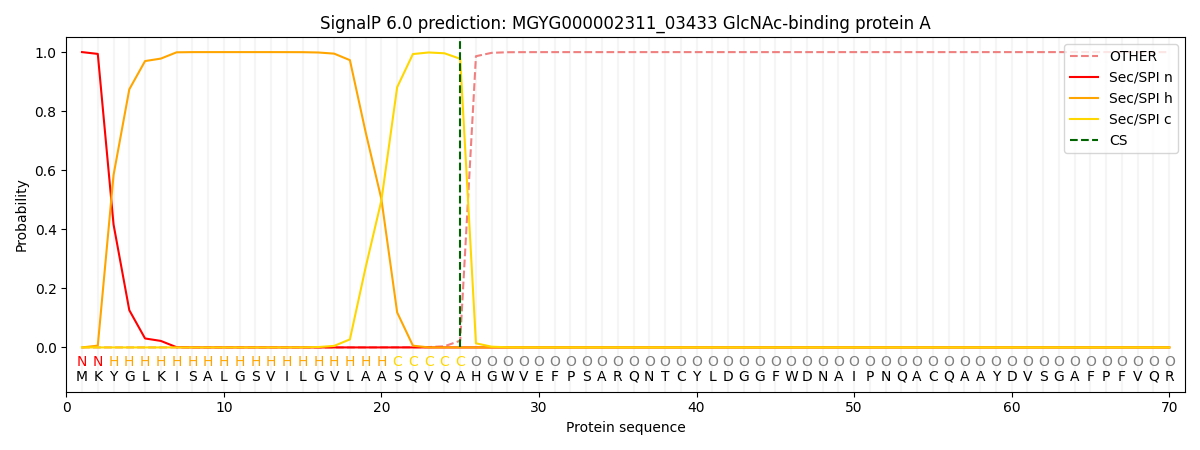
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000284 | 0.999012 | 0.000189 | 0.000173 | 0.000163 | 0.000155 |
