You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002312_01530
You are here: Home > Sequence: MGYG000002312_01530
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
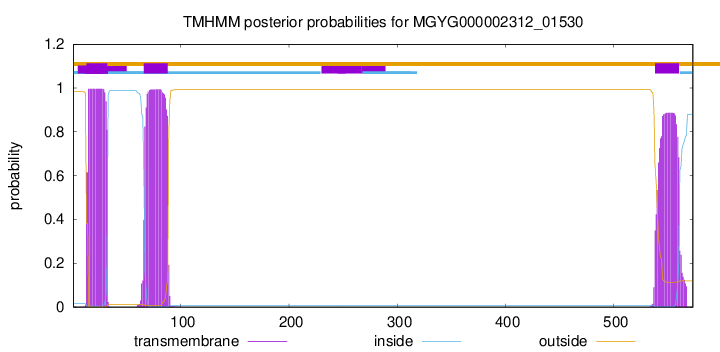
TMHMM annotations
Basic Information help
| Species | Blautia_A sp000285855 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Blautia_A; Blautia_A sp000285855 | |||||||||||
| CAZyme ID | MGYG000002312_01530 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13969; End: 15690 Strand: - | |||||||||||
Full Sequence Download help
| MDNKEVVMSK KKTIGVITGI AVGTTVVAAR IMKKKAKKTT YKVENIDSVP VRKRGFYEKY | 60 |
| IKRVCDVICA TGAIVCFSPL YICVAVLVKL KLGSPVLFTQ DRPGLIDKDG KETIFKMYKF | 120 |
| RTMTDERDEN GDLLPDEVRL TKFGKWLRNT SLDELPEAFN ILNGTMSVIG PRPQLVRDMV | 180 |
| FMTSEQRMRH TAKPGLSGLA QVNGRNAIFW ENKMEWDLKY IKKVSLHEDI KIIFKTVEKA | 240 |
| FIKQEGITEE NMATAENLGD YLLKNERITS EEYNQKQLEA KKILNRTKDK KIRDEKRITA | 300 |
| VETGAKNKKY SVLMSLYKKE KPEYLKLALD SMLNQTVIPD EIVLVEDGPL TDELYAVLNE | 360 |
| YPMLHRVKNE TNLGLGLALN VGLKECRNEL VARMDTDDCS KPDRCEKQLA RFAEKPYLAI | 420 |
| VGSHIDEFVG DISNVVSQRI VPTSSDEIYN YAKKRSAFNH PAVMYSKTAV LENDGYADLK | 480 |
| RNQDVDLFGR MQFEGYKAEN IDEALLWFRS SDELAKRRKS WQNTWSYIAT IRKFWKMGYA | 540 |
| SFYDYVMIGI AQTGMYLMPI KIQNYIYKKF LRK | 573 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 311 | 469 | 5.5e-22 | 0.9294117647058824 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd04195 | GT2_AmsE_like | 4.13e-82 | 311 | 509 | 1 | 201 | GT2_AmsE_like is involved in exopolysaccharide amylovora biosynthesis. AmsE is a glycosyltransferase involved in exopolysaccharide amylovora biosynthesis in Erwinia amylovora. Amylovara is one of the three exopolysaccharide produced by E. amylovora. Amylovara-deficient mutants are non-pathogenic. It is a subfamily of Glycosyltransferase Family GT2, which includes diverse families of glycosyltransferases with a common GT-A type structural fold, which has two tightly associated beta/alpha/beta domains that tend to form a continuous central sheet of at least eight beta-strands. These are enzymes that catalyze the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, forming glycosidic bonds. |
| pfam02397 | Bac_transf | 3.97e-71 | 62 | 241 | 1 | 180 | Bacterial sugar transferase. This Pfam family represents a conserved region from a number of different bacterial sugar transferases, involved in diverse biosynthesis pathways. |
| COG2148 | WcaJ | 1.99e-61 | 61 | 239 | 40 | 218 | Sugar transferase involved in LPS biosynthesis (colanic, teichoic acid) [Cell wall/membrane/envelope biogenesis]. |
| PRK10124 | PRK10124 | 9.32e-38 | 61 | 242 | 272 | 458 | putative UDP-glucose lipid carrier transferase; Provisional |
| PRK15204 | PRK15204 | 1.10e-28 | 59 | 246 | 277 | 474 | undecaprenyl-phosphate galactose phosphotransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARD67681.1 | 1.53e-310 | 8 | 572 | 1 | 565 |
| CCO04322.1 | 1.62e-152 | 32 | 418 | 1 | 373 |
| BAK48513.1 | 5.36e-152 | 8 | 417 | 1 | 396 |
| AOP02705.1 | 8.07e-121 | 309 | 573 | 3 | 272 |
| AOP02632.1 | 8.07e-121 | 309 | 573 | 3 | 272 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5W7L_A | 1.28e-62 | 55 | 256 | 4 | 199 | Structureof Campylobacter concisus PglC I57M/Q175M variant [Campylobacter concisus 13826],5W7L_B Structure of Campylobacter concisus PglC I57M/Q175M variant [Campylobacter concisus 13826] |
| 2Z86_A | 2.31e-07 | 323 | 421 | 388 | 488 | Crystalstructure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GlcUA and UDP [Escherichia coli],2Z86_B Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GlcUA and UDP [Escherichia coli],2Z86_C Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GlcUA and UDP [Escherichia coli],2Z86_D Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GlcUA and UDP [Escherichia coli] |
| 2Z87_A | 2.31e-07 | 323 | 421 | 387 | 487 | Crystalstructure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GalNAc and UDP [Escherichia coli],2Z87_B Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GalNAc and UDP [Escherichia coli] |
| 6P61_A | 6.52e-06 | 311 | 403 | 16 | 108 | Structureof a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197],6P61_B Structure of a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197],6P61_C Structure of a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197],6P61_D Structure of a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197] |
| 5HEA_A | 7.93e-06 | 311 | 398 | 8 | 95 | CgTstructure in hexamer [Streptococcus parasanguinis FW213],5HEA_B CgT structure in hexamer [Streptococcus parasanguinis FW213],5HEA_C CgT structure in hexamer [Streptococcus parasanguinis FW213],5HEC_A CgT structure in dimer [Streptococcus parasanguinis FW213],5HEC_B CgT structure in dimer [Streptococcus parasanguinis FW213] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0P9D0 | 1.49e-57 | 57 | 256 | 2 | 194 | Undecaprenyl phosphate N,N'-diacetylbacillosamine 1-phosphate transferase OS=Campylobacter jejuni subsp. jejuni serotype O:2 (strain ATCC 700819 / NCTC 11168) OX=192222 GN=pglC PE=1 SV=1 |
| P71062 | 3.76e-53 | 61 | 256 | 3 | 192 | Uncharacterized sugar transferase EpsL OS=Bacillus subtilis (strain 168) OX=224308 GN=epsL PE=2 SV=1 |
| Q48215 | 2.15e-43 | 309 | 573 | 2 | 267 | Uncharacterized glycosyltransferase HI_1695 OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=HI_1695 PE=3 SV=2 |
| Q03084 | 1.91e-42 | 309 | 573 | 7 | 273 | UDP-Gal:alpha-D-GlcNAc-diphosphoundecaprenol beta-1,3-galactosyltransferase OS=Escherichia coli OX=562 GN=wbbD PE=1 SV=1 |
| Q46635 | 2.04e-41 | 310 | 569 | 2 | 262 | Amylovoran biosynthesis glycosyltransferase AmsE OS=Erwinia amylovora OX=552 GN=amsE PE=3 SV=2 |
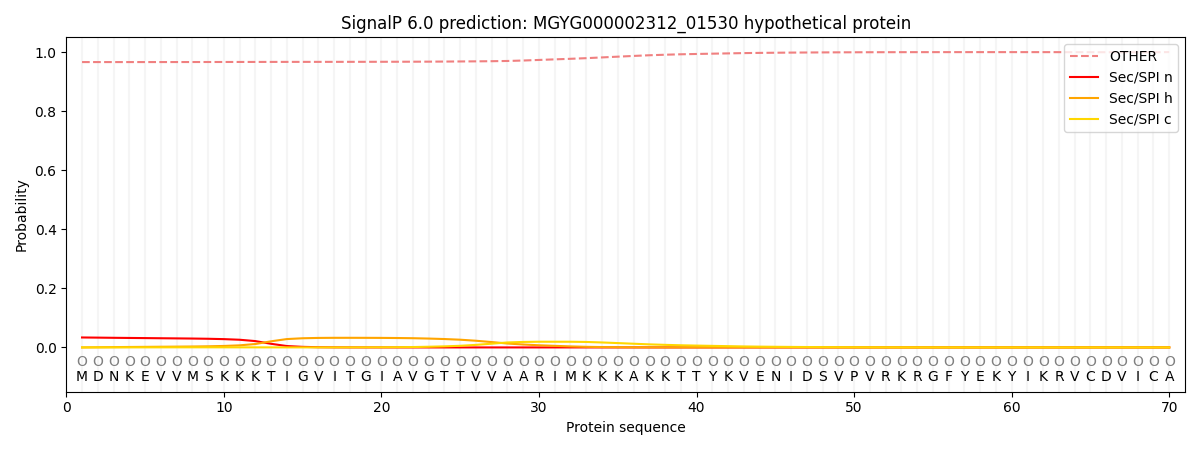
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.968067 | 0.031609 | 0.000119 | 0.000076 | 0.000058 | 0.000068 |