You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002312_03564
Basic Information
help
| Species |
Blautia_A sp000285855
|
| Lineage |
Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Blautia_A; Blautia_A sp000285855
|
| CAZyme ID |
MGYG000002312_03564
|
| CAZy Family |
GT4 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 334 |
|
39559.63 |
6.7071 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000002312 |
4168980 |
Isolate |
France |
Europe |
|
| Gene Location |
Start: 17;
End: 1021
Strand: +
|
No EC number prediction in MGYG000002312_03564.
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG0438
|
RfaB |
0.008 |
33 |
332 |
53 |
373 |
Glycosyltransferase involved in cell wall bisynthesis [Cell wall/membrane/envelope biogenesis]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
Q5UQ09
|
6.49e-156 |
1 |
330 |
266 |
595 |
Uncharacterized glycosyltransferase L193 OS=Acanthamoeba polyphaga mimivirus OX=212035 GN=MIMI_L193 PE=3 SV=1 |
This protein is predicted as OTHER
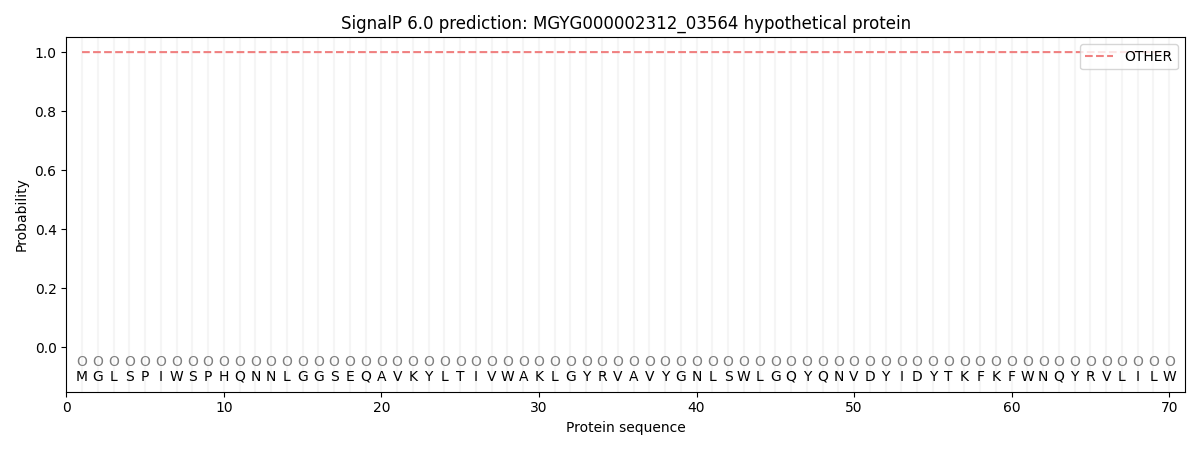
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.999926
|
0.000072
|
0.000005
|
0.000000
|
0.000000
|
0.000003
|
There is no transmembrane helices in MGYG000002312_03564.

