You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002331_01462
You are here: Home > Sequence: MGYG000002331_01462
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Vibrio parahaemolyticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Vibrionaceae; Vibrio; Vibrio parahaemolyticus | |||||||||||
| CAZyme ID | MGYG000002331_01462 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | putative L,D-transpeptidase YcfS | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1627261; End: 1628187 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK10260 | PRK10260 | 4.80e-101 | 29 | 267 | 24 | 262 | L,D-transpeptidase; Provisional |
| PRK10190 | PRK10190 | 4.37e-89 | 29 | 308 | 21 | 305 | L,D-transpeptidase; Provisional |
| COG1376 | ErfK | 5.15e-52 | 28 | 242 | 22 | 232 | Lipoprotein-anchoring transpeptidase ErfK/SrfK [Cell wall/membrane/envelope biogenesis]. |
| cd16913 | YkuD_like | 3.88e-40 | 104 | 239 | 2 | 121 | L,D-transpeptidases/carboxypeptidases similar to Bacillus YkuD. Members of the YkuD-like family of proteins are found in a range of bacteria. The best studied member Bacillus YkuD has been shown to act as an L,D-transpeptidase that gives rise to an alternative pathway for peptidoglycan cross-linking. Another member Helicobacter pylori Csd6 functions as an L,D-carboxypeptidase and regulates helical cell shape and motility. The conserved region contains a conserved histidine and cysteine, with the cysteine thought to be an active site residue. |
| pfam17969 | Ldt_C | 5.78e-24 | 242 | 305 | 1 | 67 | L,D-transpeptidase C-terminal domain. This is the C-terminal domain found in d-transpeptidases (Ldt) homologues from E.coli. Three of these enzymes (YbiS, ErfK, YcfS) have been shown to cross-link Braun's lipoprotein to the peptidoglycan (PG), while the other two (YnhG, YcbB) form direct meso-diaminopimelate (DAP-DAP, or 3-3) cross-links within the PG. Family members include erfK (ldtA), ybiS (ldtB), ycfS (ldtC), and ynhG (ldtE). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUY00823.1 | 3.47e-149 | 29 | 308 | 20 | 299 |
| QPC55488.1 | 3.47e-149 | 29 | 308 | 20 | 299 |
| QTV10206.1 | 3.47e-149 | 29 | 308 | 20 | 299 |
| QLK92159.1 | 3.47e-149 | 29 | 308 | 20 | 299 |
| QTV04013.1 | 3.47e-149 | 29 | 308 | 20 | 299 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4LZH_A | 1.59e-77 | 27 | 267 | 1 | 241 | ChainA, L,D-transpeptidase [Klebsiella pneumoniae subsp. pneumoniae MGH 78578] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P75954 | 1.97e-94 | 29 | 305 | 28 | 310 | Probable L,D-transpeptidase YcfS OS=Escherichia coli (strain K12) OX=83333 GN=ycfS PE=1 SV=1 |
| P76193 | 7.79e-87 | 18 | 262 | 7 | 256 | Probable L,D-transpeptidase YnhG OS=Escherichia coli (strain K12) OX=83333 GN=ynhG PE=1 SV=1 |
| P0AAX8 | 6.77e-84 | 8 | 267 | 1 | 262 | Probable L,D-transpeptidase YbiS OS=Escherichia coli (strain K12) OX=83333 GN=ybiS PE=1 SV=1 |
| P0AAX9 | 6.77e-84 | 8 | 267 | 1 | 262 | Probable L,D-transpeptidase YbiS OS=Escherichia coli O6:H1 (strain CFT073 / ATCC 700928 / UPEC) OX=199310 GN=ybiS PE=3 SV=1 |
| P0AAY0 | 6.77e-84 | 8 | 267 | 1 | 262 | Probable L,D-transpeptidase YbiS OS=Escherichia coli O157:H7 OX=83334 GN=ybiS PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
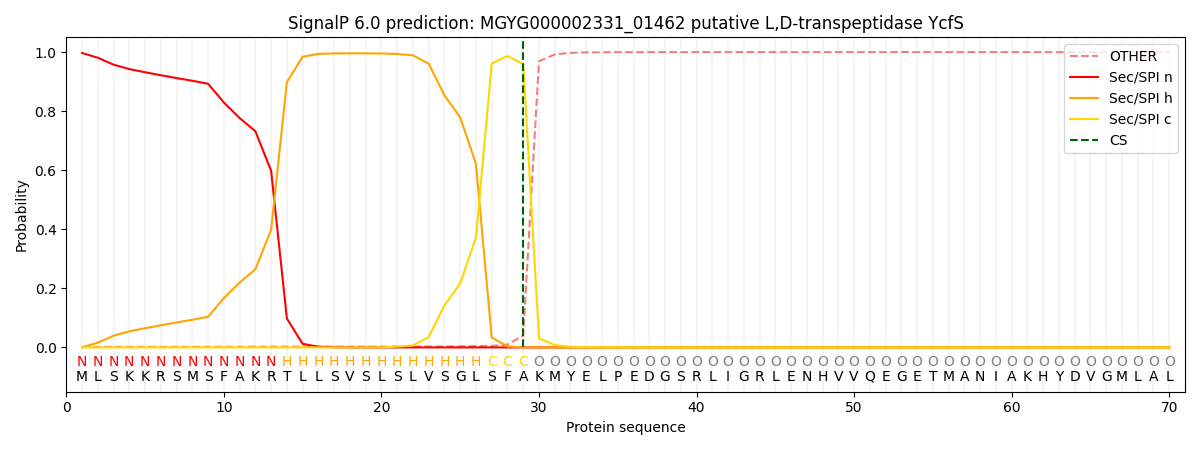
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003199 | 0.994399 | 0.001593 | 0.000303 | 0.000257 | 0.000209 |
