You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002334_00431
You are here: Home > Sequence: MGYG000002334_00431
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
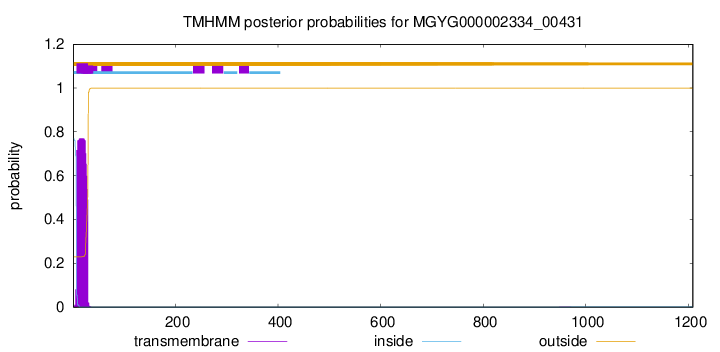
TMHMM annotations
Basic Information help
| Species | Bifidobacterium thermophilum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium thermophilum | |||||||||||
| CAZyme ID | MGYG000002334_00431 | |||||||||||
| CAZy Family | CBM26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 560153; End: 563782 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM26 | 222 | 285 | 1.6e-19 | 0.8933333333333333 |
| CBM26 | 130 | 193 | 1.3e-18 | 0.8933333333333333 |
| CBM26 | 39 | 108 | 1.5e-18 | 0.9333333333333333 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16738 | CBM26 | 2.05e-19 | 222 | 279 | 1 | 60 | Starch-binding module 26. CBM26 is a carbohydrate-binding module that binds starch. |
| pfam16738 | CBM26 | 6.12e-19 | 131 | 195 | 1 | 68 | Starch-binding module 26. CBM26 is a carbohydrate-binding module that binds starch. |
| pfam16738 | CBM26 | 5.81e-18 | 40 | 104 | 1 | 68 | Starch-binding module 26. CBM26 is a carbohydrate-binding module that binds starch. |
| cd13444 | CDI_toxin_EC869_like | 1.66e-17 | 1058 | 1207 | 3 | 139 | Zn-dependent DNAse of the contact-dependent growth inhibition (CDI) system of Escherichia coli EC869, and related proteins. CDI toxins are expressed by gram-negative bacteria as part of a mechanism to inhibit the growth of neighboring bacteria. This model represents the C-terminal toxin domain of CdiA effector proteins. CdiA secretion is dependent on the outer membrane protein CdiB. Upon binding to a receptor on the surface of target bacteria, the CDI toxin is delivered. A wide variety of C-terminal toxin domains appear to exist; this particular example from Escherichia coli EC869 and other bacteria appears to function as a Zn2+-dependent DNAse degrading the genome of target cells. |
| cd00198 | vWFA | 3.21e-15 | 580 | 734 | 4 | 160 | Von Willebrand factor type A (vWA) domain was originally found in the blood coagulation protein von Willebrand factor (vWF). Typically, the vWA domain is made up of approximately 200 amino acid residues folded into a classic a/b para-rossmann type of fold. The vWA domain, since its discovery, has drawn great interest because of its widespread occurrence and its involvement in a wide variety of important cellular functions. These include basal membrane formation, cell migration, cell differentiation, adhesion, haemostasis, signaling, chromosomal stability, malignant transformation and in immune defenses In integrins these domains form heterodimers while in vWF it forms multimers. There are different interaction surfaces of this domain as seen by the various molecules it complexes with. Ligand binding in most cases is mediated by the presence of a metal ion dependent adhesion site termed as the MIDAS motif that is a characteristic feature of most, if not all A domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGH40717.1 | 0.0 | 1 | 1209 | 1 | 1209 |
| CBK93270.1 | 3.22e-218 | 307 | 1209 | 338 | 1237 |
| QEH68420.1 | 8.64e-60 | 37 | 308 | 886 | 1161 |
| QUL54870.1 | 3.12e-55 | 307 | 837 | 376 | 903 |
| CDZ23468.1 | 4.19e-49 | 330 | 803 | 369 | 844 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2C3G_A | 7.49e-11 | 125 | 217 | 3 | 97 | ChainA, ALPHA-AMYLASE G-6 [Halalkalibacterium halodurans C-125],2C3H_A Chain A, ALPHA-AMYLASE G-6 [Halalkalibacterium halodurans],2C3H_B Chain B, ALPHA-AMYLASE G-6 [Halalkalibacterium halodurans],2C3H_C Chain C, ALPHA-AMYLASE G-6 [Halalkalibacterium halodurans],2C3H_D Chain D, ALPHA-AMYLASE G-6 [Halalkalibacterium halodurans],2C3H_E Chain E, ALPHA-AMYLASE G-6 [Halalkalibacterium halodurans],2C3H_F Chain F, ALPHA-AMYLASE G-6 [Halalkalibacterium halodurans],2C3H_G Chain G, ALPHA-AMYLASE G-6 [Halalkalibacterium halodurans],2C3H_H Chain H, ALPHA-AMYLASE G-6 [Halalkalibacterium halodurans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P30269 | 1.40e-06 | 33 | 166 | 815 | 973 | Alpha-amylase OS=Butyrivibrio fibrisolvens OX=831 GN=amyA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000230 | 0.999069 | 0.000197 | 0.000180 | 0.000161 | 0.000144 |