You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002335_04017
You are here: Home > Sequence: MGYG000002335_04017
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
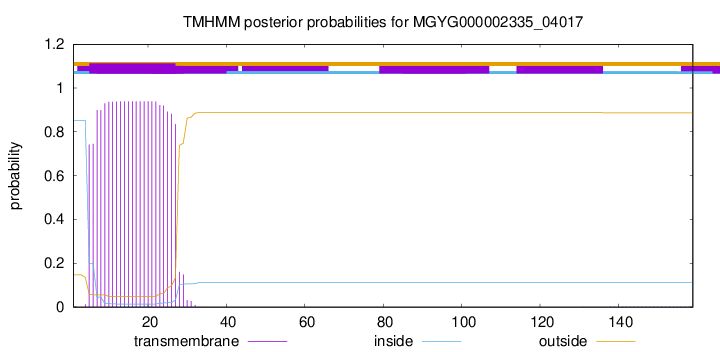
TMHMM annotations
Basic Information help
| Species | Yersinia enterocolitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Yersinia; Yersinia enterocolitica | |||||||||||
| CAZyme ID | MGYG000002335_04017 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4340152; End: 4340631 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 28 | 145 | 5.6e-23 | 0.9274193548387096 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.53e-17 | 26 | 145 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam17132 | Glyco_hydro_106 | 1.30e-07 | 26 | 129 | 175 | 282 | alpha-L-rhamnosidase. |
| cd00057 | FA58C | 5.66e-07 | 27 | 142 | 14 | 137 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QBP97459.1 | 4.46e-109 | 1 | 159 | 1 | 159 |
| VEA97796.1 | 4.46e-109 | 1 | 159 | 1 | 159 |
| AOF13314.1 | 4.46e-109 | 1 | 159 | 1 | 159 |
| VEF82428.1 | 4.46e-109 | 1 | 159 | 1 | 159 |
| CBY29402.1 | 4.46e-109 | 1 | 159 | 1 | 159 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2JD9_A | 2.18e-96 | 20 | 159 | 6 | 145 | Structureof a pectin binding carbohydrate binding module determined in an orthorhombic crystal form. [Yersinia enterocolitica],2JDA_A Structure of a pectin binding carbohydrate binding module determined in an monoclinic crystal form. [Yersinia enterocolitica],2JDA_B Structure of a pectin binding carbohydrate binding module determined in an monoclinic crystal form. [Yersinia enterocolitica] |
| 7D29_A | 9.45e-17 | 24 | 147 | 11 | 131 | CBM32of AlyQ [Persicobacter sp. CCB-QB2],7D2A_A CBM32 of AlyQ in complex with 4,5-unsaturated mannuronic acid [Persicobacter sp. CCB-QB2] |
| 5XNR_A | 1.76e-15 | 24 | 147 | 11 | 131 | TruncatedAlyQ with CBM32 and alginate lyase domains [Persicobacter sp. CCB-QB2] |
| 2VC9_A | 1.07e-08 | 25 | 154 | 8 | 137 | Family89 Glycoside Hydrolase from Clostridium perfringens in complex with 2-acetamido-1,2-dideoxynojirmycin [Clostridium perfringens],2VCA_A Family 89 glycoside hydrolase from Clostridium perfringens in complex with beta-N-acetyl-D-glucosamine [Clostridium perfringens],2VCB_A Family 89 Glycoside Hydrolase from Clostridium perfringens in complex with PUGNAc [Clostridium perfringens],2VCC_A Family 89 Glycoside Hydrolase from Clostridium perfringens [Clostridium perfringens] |
| 7MFK_A | 1.07e-08 | 25 | 154 | 16 | 145 | ChainA, Alpha-N-acetylglucosaminidase family protein [Clostridium perfringens ATCC 13124],7MFL_A Chain A, Alpha-N-acetylglucosaminidase family protein [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
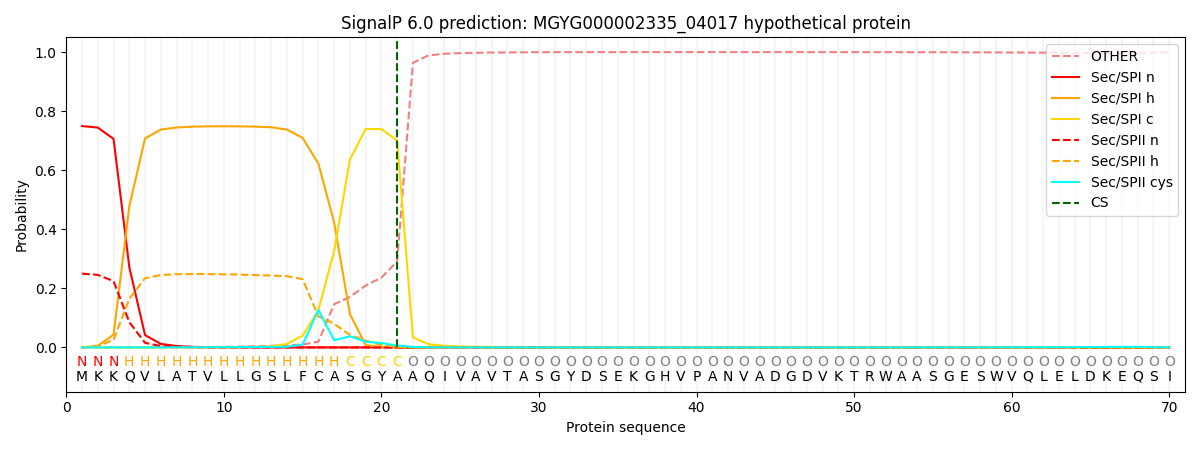
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001574 | 0.738473 | 0.258875 | 0.000492 | 0.000297 | 0.000264 |