You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002355_00303
You are here: Home > Sequence: MGYG000002355_00303
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridium_F botulinum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium_F; Clostridium_F botulinum | |||||||||||
| CAZyme ID | MGYG000002355_00303 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 275038; End: 277323 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 2.53e-55 | 33 | 705 | 33 | 531 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231 | PMT_2 | 4.26e-45 | 66 | 224 | 1 | 160 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG1928 | PMT1 | 7.68e-04 | 67 | 135 | 90 | 159 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| pfam06800 | Sugar_transport | 0.001 | 70 | 111 | 157 | 197 | Sugar transport protein. This is a family of bacterial sugar transporters approximately 300 residues long. Members include glucose uptake proteins, ribose transport proteins, and several putative and hypothetical membrane proteins probably involved in sugar transport across bacterial membranes. These members are transmembrane proteins which are usually 5+5 duplications. This model recognizes a set of five TMs, |
| COG1287 | Stt3 | 0.002 | 65 | 224 | 85 | 212 | Asparagine N-glycosylation enzyme, membrane subunit Stt3 [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUB37647.1 | 3.31e-66 | 16 | 700 | 210 | 780 |
| QHU89858.1 | 5.12e-66 | 31 | 701 | 293 | 890 |
| QJU08762.1 | 4.80e-60 | 31 | 701 | 292 | 896 |
| QHU92512.1 | 6.02e-60 | 31 | 701 | 291 | 887 |
| QJU05422.1 | 8.50e-60 | 31 | 701 | 293 | 891 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EZM_A | 3.36e-07 | 66 | 229 | 87 | 255 | CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34575 | 6.70e-121 | 10 | 714 | 9 | 678 | Putative mannosyltransferase YkcB OS=Bacillus subtilis (strain 168) OX=224308 GN=ykcB PE=3 SV=2 |
| P37483 | 2.29e-115 | 5 | 708 | 2 | 659 | Putative mannosyltransferase YycA OS=Bacillus subtilis (strain 168) OX=224308 GN=yycA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
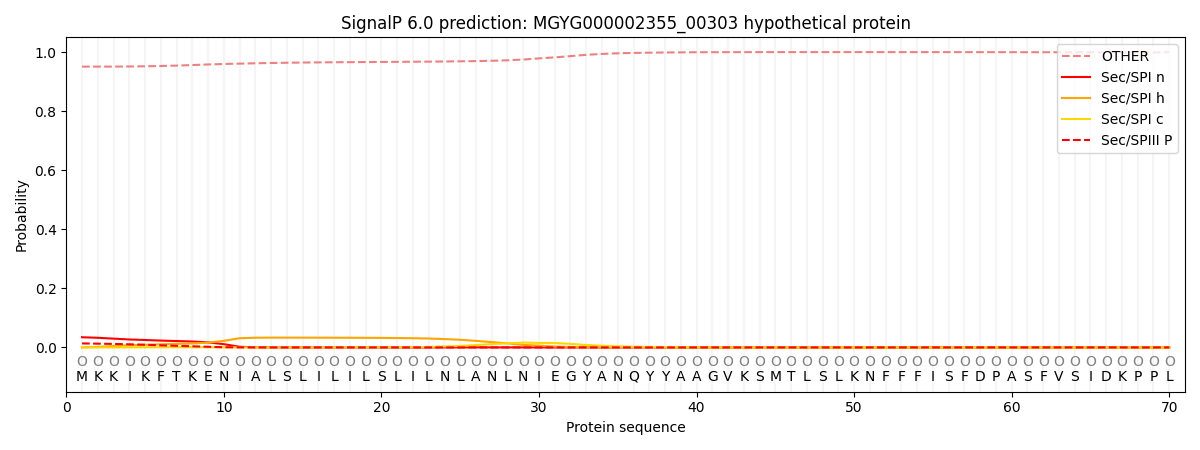
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.952427 | 0.031933 | 0.001460 | 0.000161 | 0.000119 | 0.013927 |
TMHMM Annotations download full data without filtering help
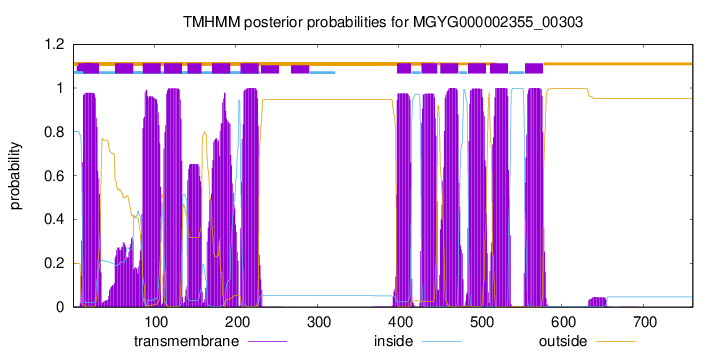
| start | end |
|---|---|
| 13 | 32 |
| 52 | 74 |
| 86 | 108 |
| 112 | 134 |
| 141 | 158 |
| 171 | 193 |
| 206 | 228 |
| 398 | 415 |
| 428 | 447 |
| 451 | 473 |
| 485 | 507 |
| 512 | 534 |
| 555 | 577 |
